Objective: To write a user defined function that automates the wrangling process for multiple .csv files.
I have 3 .csv filesnamely, Finland_caucasian.csv, Mexico_hispanic.csv & Jamaica_black.csv.
Using Finland_caucasian.csv as an example, the dataframe looks like this :
date <- rep('1999',9)
cityID <- c(1001,1001,1001,1002,1002,1002,1005,1005,1005)
country <- rep('FIN'9)
name <- rep('caucasian',9)
prov_code <- c(1,1,1,2,2,3,3,2,3)
class <- c(1,2,5,1,2,5,1,2,5)
gender <- c('M','M','F','F','F','M','F','M','M')
savings_month1 <-c(100.12,105.11,109.11,94.13,34.00,201.91,154.39,212.76,233.48)
savings_month2 <-c(110.15,115.12,193.21,194.13,334.50,21.13,164.19,292.67,235.85)
savings_month3 <-c(109.15,159.12,199.22,199.18,339.59,291.95,169.94,299.78,233.22)
data<- data.frame(date,cityID,name,country,prov_code,class,gender,savings_month1,savings_month2,savings_month3)
data
The raw wrangling process (without a user-defined function) looks like this :
# template for the user-defined function- should import csv files in a folder
#############################################################################
data_wrangler <- function(input_multi_csvfiles){
# import each csv file
for(files in filenames){
# wrangle process
}
}
# test code
data_wrangler(files in folder path)
################################################################################################
# code for the wrangling process
library(dplyr)
#Step I: import all three files in R
data = read.csv("filepath//file_name.csv") # 'Finland_caucasian.csv','Mexico_hispanic.csv' and 'Jamaica_black.csv'
#Step II : select the desired column names
df <- data%>%select(date:gender,savings_month1:savings_month3) # I used column index `:` because the other csv files might have undesired columns in between.
#Step III: check the unique prov_code, country,name, class
print(unique(df$prov_code))
print(unique(df$country))
print(unique(df$name))
print(unique(df$class))
#Step IV: filter the df based on 'class'
# Here, I will like to automate to reflect the unique countries ( FIN, MEX and JAM) instead of manually entering the countries three times, likewise for the name column- caucasian, hispanic and black
country_name_df <-subset(df,country == 'FIN' & name == 'caucasian' & class >= 1 & class <= 5) # in the other csv files class ranges from 0 to 8, but I want to filter for ONLY class ranging from 1 to 5
#Step V: Paste filtered class on each of the column names
df10 <- filter(country_name_df,class == 1)
#paste the filtered class condition to the column names
colnames(df10) <- paste('1',colnames(df10),sep = '_')
df11 <- filter(country_name_df,class == 2)
#paste the filtered class condition to the column names
colnames(df11) <- paste('2',colnames(df11),sep = '_')
df12 <- filter(country_name_df,class == 5)
#paste the filtered class condition to the column names
colnames(df12) <- paste('5',colnames(df12),sep = '_')
#Step VI: column bind or merge df10, df11 and df12
df15 <- cbind(df10,df11,df12)
#check for content of class column of df15
unique(df15$`5_prov_code`)
#Step VII: final wrangle for the dataframe
df20<- df15%>%select(`1_date`:`1_gender`)
df30 <-select(df15,contains("month")) # select all columns containing string 'month' example '1_savings_month1', '3_savings_month2' etc
target <- df15$`1_prov_code`
final_df <- cbind(df20,df30,target)
#Step VIII: export each of the final_df as csv files
# wrangled_Finland caucasian.csv, wrangled_Mexico hispanic.csv,wrangled_Jamaica black.csv
write.csv(final_df,'wrangled_FIN caucasian.csv',row.names = FALSE)
I am open to other ideas. Thanks
Update for Akrun
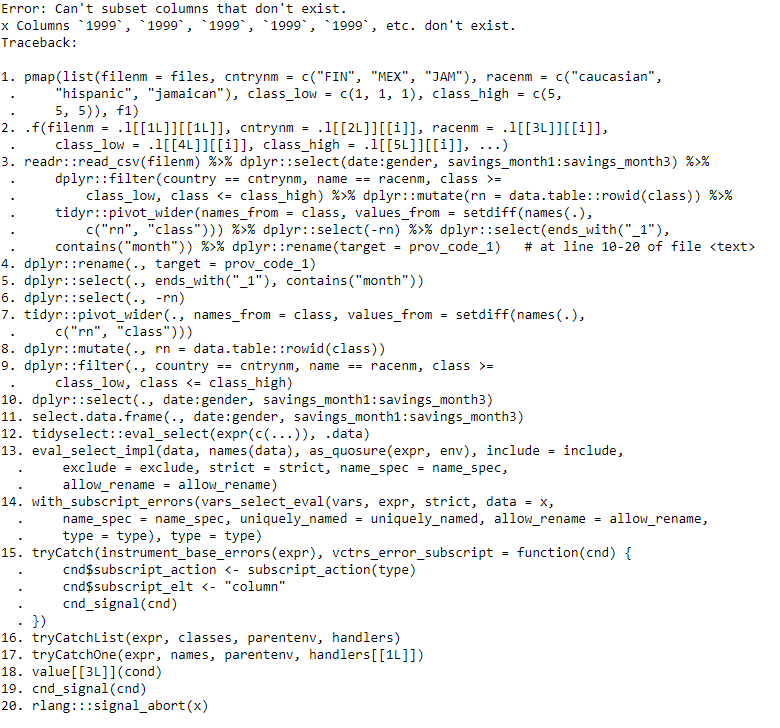
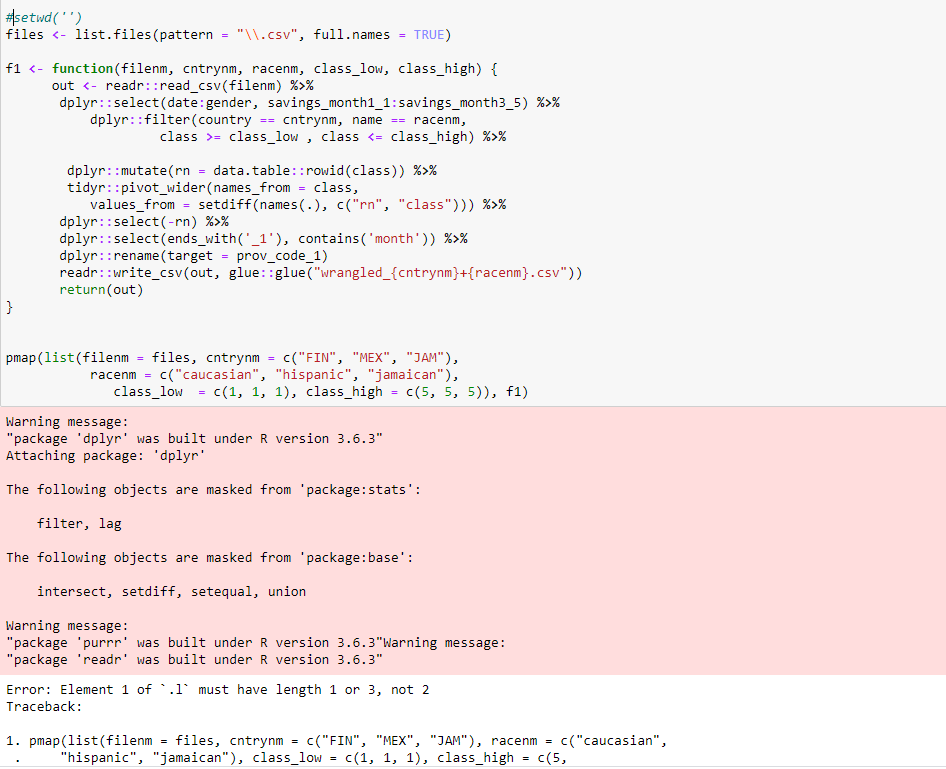
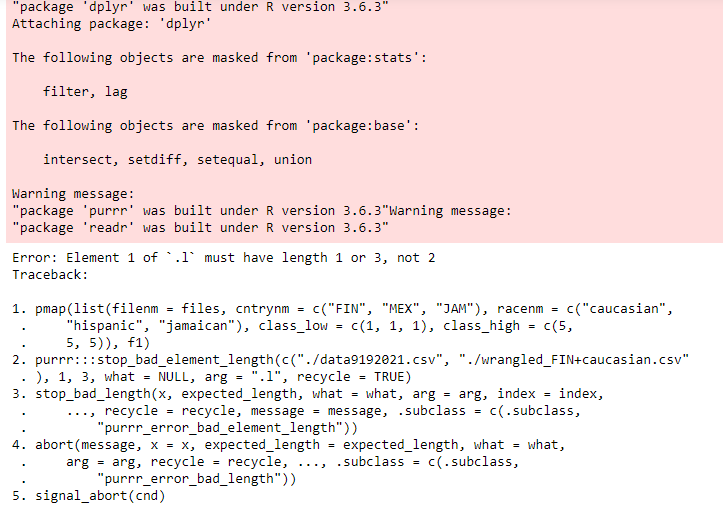
I ran your code (see below) .
error i got
Update 2: sequence of column names after gender_1 should look like this savings_month1_1, savings_month1_2,savings_month1_5, savings_month2_1,savings_month2_2,savings_month2_5,savings_month3_1,savings_month3_2,savings_month3_5
Update 3: error after changing savings_month1:savings_month3
CodePudding user response:
If we have 3 files in the working directory, read those in a list with map, do the select and filter step and use either group_by on 'class' or split into a list of data.frames
library(dplyr)
library(purrr)
library(readr)
files <- list.files(pattern = "\\.csv", full.names = TRUE)
df_list <- map(files, ~ read_csv(.x) %>%
select(date:gender, savings_month1:savings_month3) %>%
filter(country == 'FIN', name == 'caucasian',
class >= 1 , class <= 5) %>%
group_by(class))
if we need a function with parameters
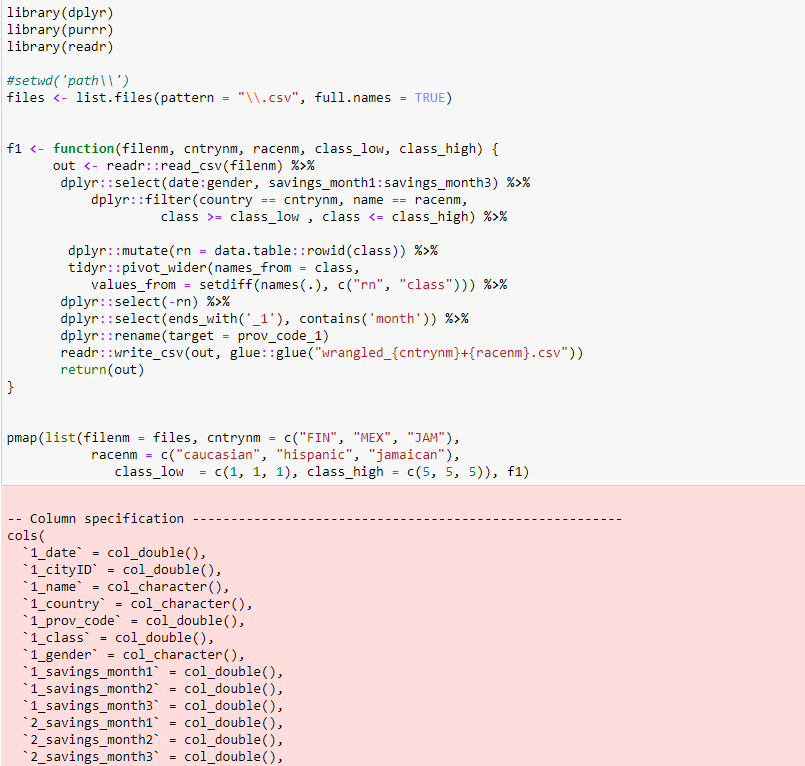
f1 <- function(filenm, cntrynm, racenm, class_low, class_high) {
out <- readr::read_csv(filenm) %>%
dplyr::select(date:gender, savings_month1_1:savings_month3_5) %>%
dplyr::filter(country == cntrynm, name == racenm,
class >= class_low , class <= class_high) %>%
dplyr::mutate(rn = data.table::rowid(class)) %>%
tidyr::pivot_wider(names_from = class,
values_from = setdiff(names(.), c("rn", "class"))) %>%
dplyr::select(-rn) %>%
dplyr::select(ends_with('_1'), contains('month')) %>%
dplyr::rename(target = prov_code_1)
readr::write_csv(out, glue::glue("wrangled_{cntrynm} {racenm}.csv"))
return(out)
}
Then, we apply the function with the parameters in pmap
pmap(list(filenm = files, cntrynm = c("FIN", "MEX", "JAM"),
racenm = c("caucasian", "hispanic", "jamaican"),
class_low = c(1, 1, 1), class_high = c(5, 5, 5)), f1)