I am trying to make multiple stacked barplots with a phyloseq object (containing OTU, TAX, and Metadata) in which the order of taxa on the barplot stays the same, but on each graph a particular taxa is colorized while the rest are grey. I written a rough draft of code using a simple data frame, but i'm not sure how to apply this to a phyloseq object.
Here is the code i've come up with:
##good_phylum being the specfic taxa i want colorized
good_phylum <- c("p__Firmicutes")
## pig_df contains taxa information and relative abundance
pig_df[!(pig_df$Phylum %in% good_phylum),]$Phylum <- "OTHER"
##organizing color palette so Firmicutes is colored blue, the rest are grey
colors <- rep("#0000", length(unique(pig_df$Phylum)))
colors[match('p__Firmicutes', sort(unique(pig_df$Phylum)))] <- 'blue'
##Plot stacked bar plot
ggplot2(pig_df) geom_bar(position="stack",
stat="Phylum") scale_fill_manual(values = colors)
theme(legend.text = element_text(size = 14))
Sample data:
structure(list(Phylum = c("p__Actinobacteria", "p__Bacteroidetes",
"p__Firmicutes", "p__Lentisphaerae", "p__Proteobacteria", "p__Spirochaetes"
), AL.5 = c(5.54534, 44.05287, 37.01594, 0, 2.71389, 10.67196
), AL.4 = c(0, 40.61791, 29.41689, 0, 0.39118, 29.57402), AL.3 = c(0.51442,
39.55305, 35.79623, 0, 2.79092, 21.34539), AL.2 = c(2.97639,
53.8286, 25.08614, 0, 8.70794, 9.40094), AL.1 = c(3.35874, 39.20605,
25.74435, 0.30047, 9.23324, 22.15715), LF.5 = c(0.40971, 3.95695,
17.63298, 0.06856, 76.56688, 1.36492), LF.4 = c(2.7231, 12.9073,
81.70363, 0.09188, 1.12626, 1.44782), LF.3 = c(9.58431, 7.14942,
56.77299, 0, 20.15845, 6.33483), LF.2 = c(0.26317, 17.63049,
27.01207, 0, 53.73687, 1.3574), LF.1 = c(5.48864, 36.46061, 41.32865,
0, 1.71807, 15.00404)), class = "data.frame", row.names = c(NA,
-6L))
Like i said Ive written the above to work with a simple data frame, but i would like to work this into my phyloseq workflow.
Thanks in advance!
*Edit: updated with a minimal sample data frame, and code was updated to work with said df
CodePudding user response:
Not 100% percent about your desired result, but as far as I get it one option would be:
- Make use of a named vector of colors.
- To get your bars colored you have to map on the
fillaesthetic. - As you mentioned that you want to make multiple charts I would suggest to make use of a plotting function:
Note: I dropped stat="Phylum" as I wasn't able to figure out from which package this stat comes from.
library(ggplot2)
plot_phylum <- function(pig_df, good_phylum) {
colors <- c("grey", "blue")
names(colors) < c("FALSE", "TRUE")
labels <- c("OTHER", good_phylum)
names(labels) < c("FALSE", "TRUE")
##Plot stacked bar plot
ggplot(pig_df, aes(x = factor(1), fill = Phylum %in% good_phylum))
geom_bar(position="stack")
scale_fill_manual(values = colors, labels = labels, name = NULL)
theme(legend.text = element_text(size = 14))
}
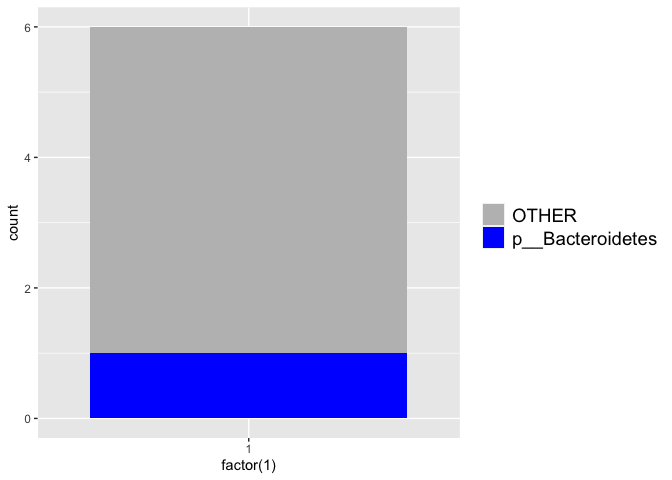
plot_phylum(pig_df, "p__Firmicutes")
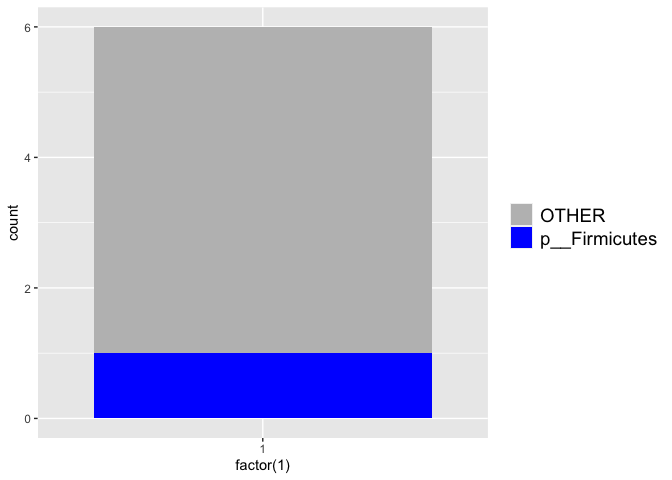
plot_phylum(pig_df, "p__Bacteroidetes")