I was trying to plot a line using ggplot but because the x values (days) are not even spaced I do not get a solid line. I tried instead of using geom_line() to use geom_smooth() but I get the lines with confidence intervals.
My data looks like this:
#df
treatment day1 day2 day4 day6 day9
1 12,7 11,1 8,7 7,4 5,9
2 13,4 11,3 8,6 6,9 5,3
3 11,3 10,5 8,9 7,6 6,2
Is it any other geometric expression or way to arrange the data to plot a solid continuous line? Thanks in advance
CodePudding user response:
I'm not sure what you want exactly but it can look something like the following:
library(tidyverse)
data <- tibble(treatment = 1:3,
day1 = c(12.7, 13.4, 11.3),
day2 = c(11.1, 11.3, 10.5),
day4 = c(8.7, 8.6, 8.9),
day6 = c(7.4, 6.9, 7.6),
day9 = c(5.9, 5.3 ,6.2))
data %>%
pivot_longer(-treatment, names_to = "days", names_prefix = "day") %>%
mutate(days = as.integer(days),
treatment = as.character(treatment)) %>%
ggplot(aes(days, value, color = treatment))
geom_line()
If it's not the case it can be helpful if you specify the graph you hope to get (x and y variable) and the code you have run.
CodePudding user response:
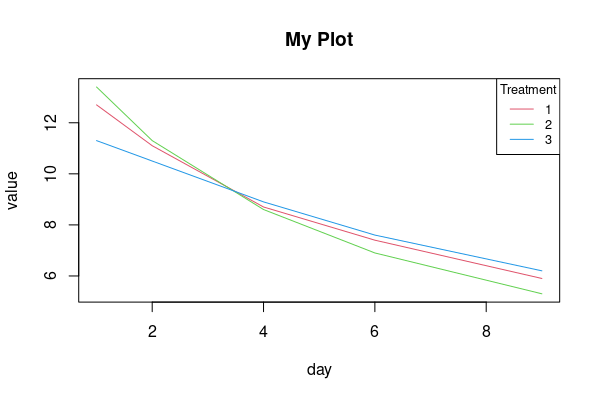
You could use melt from reshape2 package and base graphics.
d1 <- reshape2::melt(d0, id=1, variable.name = "day") |>
transform(day=as.numeric(gsub('day', '', day)))
plot(value ~ day, d1, type='n', main='My Plot')
by(d1, d1$treatment, \(x) lines(value ~ day, x, col=x[1, 1] 1))
legend('topright', legend=1:3, title='Treatment', lty=1, col=2:4, cex=.8)
Data
d0 <- read.table(header=T, text='treatment day1 day2 day4 day6 day9
1 12,7 11,1 8,7 7,4 5,9
2 13,4 11,3 8,6 6,9 5,3
3 11,3 10,5 8,9 7,6 6,2', dec=',')