I have multiple .csv files (currently 4 but will be variable in the future) and am trying to import them into R (into a single data frame).
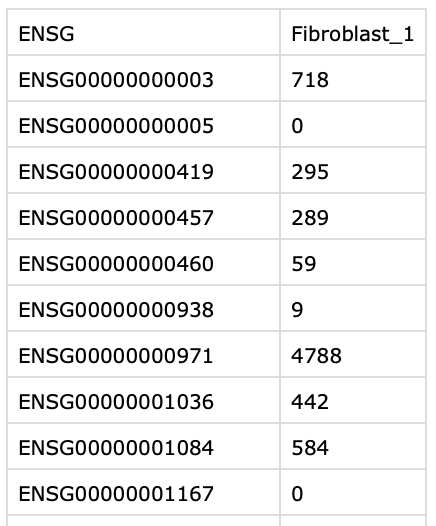
I extracted the file names and used that in lapply to load them as a list of lists and defined the row names. Each file has a row names column and a column with the data and looks like:
each separate file
list of lists in R after lapply
count_files <- list.files()
count_lists <- lapply(count_files, read.csv, sep=",", header=TRUE, row.names="ENSG")
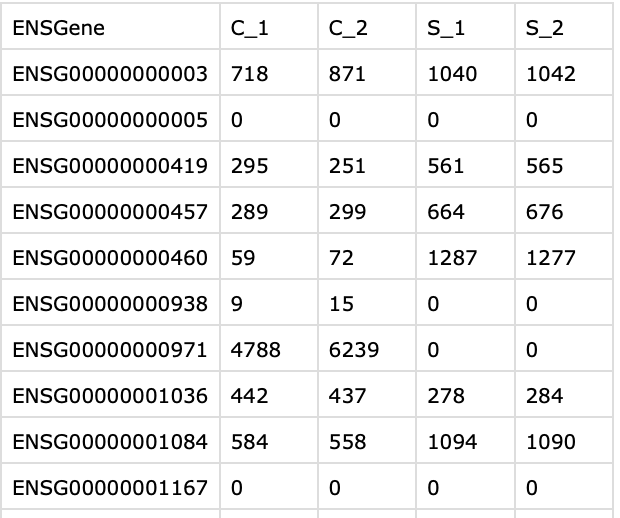
What I need is for the final data frame in R to look like:
final data frame in R
CodePudding user response:
You may try using merge in Reduce and join the dataframes by rownames.
result <- Reduce(function(x, y) merge(x, y, by = 'row.names', all = TRUE), count_lists)
result[is.na(result)] <- 0
result
CodePudding user response:
We may do this in tidyverse
library(dplyr)
library(purrr)
map(count_lists, ~ .x %>%
rownames_to_column('rn')) %>%
reduce(full_join, by = 'rn') %>%
mutate(across(everything(), replace_na, 0))