I would like to add annotation info(anno) to the matching samples in the logtpm2 file.
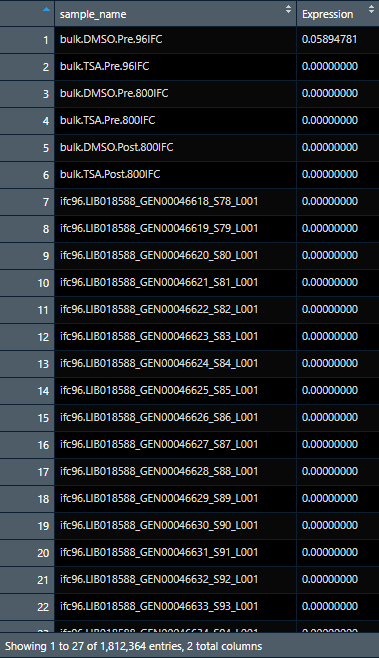
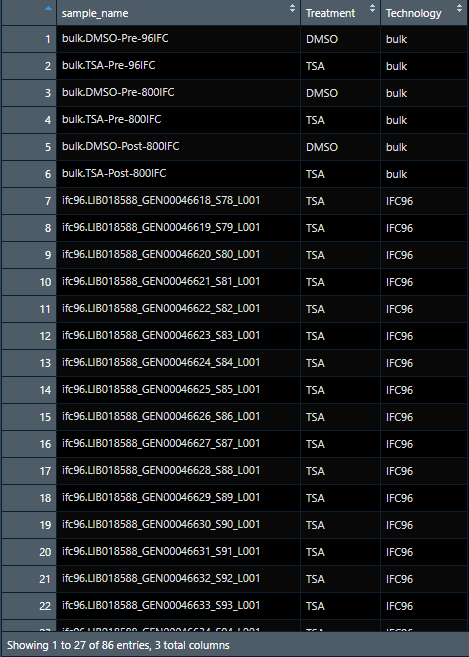
for example, first picture here represents logtpm2 file (1,812,364 entries with expression data), and I would like to add 2 columns from the annotation data set (anno) which contains Treatment, and Technology columns. How do I match the logtpm2 data with the corresponding data sample? Any help will be appreciated. Thanks.
The ideal data will have 1,812,364 rows with 3 columns in the end: Expression, Treatment, and technology
data sample for logtpm2
sample_name Expression
1 bulk.DMSO.Pre.96IFC 0.05894781
2 bulk.TSA.Pre.96IFC 0.00000000
3 bulk.DMSO.Pre.800IFC 0.00000000
4 bulk.TSA.Pre.800IFC 0.00000000
5 bulk.DMSO.Post.800IFC 0.00000000
6 bulk.TSA.Post.800IFC 0.00000000
7 ifc96.LIB018588_GEN00046618_S78_L001 0.00000000
8 ifc96.LIB018588_GEN00046619_S79_L001 0.00000000
9 ifc96.LIB018588_GEN00046620_S80_L001 0.00000000
10 ifc96.LIB018588_GEN00046621_S81_L001 0.00000000
11 ifc96.LIB018588_GEN00046622_S82_L001 0.00000000
12 ifc96.LIB018588_GEN00046623_S83_L001 0.00000000
13 ifc96.LIB018588_GEN00046624_S84_L001 0.00000000
14 ifc96.LIB018588_GEN00046625_S85_L001 0.00000000
15 ifc96.LIB018588_GEN00046626_S86_L001 0.00000000
16 ifc96.LIB018588_GEN00046627_S87_L001 0.00000000
17 ifc96.LIB018588_GEN00046628_S88_L001 0.00000000
18 ifc96.LIB018588_GEN00046629_S89_L001 0.00000000
19 ifc96.LIB018588_GEN00046630_S90_L001 0.00000000
20 ifc96.LIB018588_GEN00046631_S91_L001 0.00000000
21 ifc96.LIB018588_GEN00046632_S92_L001 0.00000000
22 ifc96.LIB018588_GEN00046633_S93_L001 0.00000000
23 ifc96.LIB018588_GEN00046634_S94_L001 0.00000000
24 ifc96.LIB018588_GEN00046635_S95_L001 0.00000000
25 ifc96.LIB018588_GEN00046636_S96_L001 0.00000000
26 ifc96.LIB018589_GEN00046637_S97_L002 0.00000000
27 ifc96.LIB018589_GEN00046638_S98_L002 0.00000000
28 ifc96.LIB018589_GEN00046639_S99_L002 0.00000000
29 ifc96.LIB018589_GEN00046640_S100_L002 0.00000000
30 ifc96.LIB018589_GEN00046641_S101_L002 0.00000000
31 ifc96.LIB018589_GEN00046642_S102_L002 0.00000000
32 ifc96.LIB018589_GEN00046643_S103_L002 0.00000000
33 ifc96.LIB018589_GEN00046644_S104_L002 0.00000000
34 ifc96.LIB018589_GEN00046645_S105_L002 0.00000000
35 ifc96.LIB018589_GEN00046646_S106_L002 0.00000000
36 ifc96.LIB018589_GEN00046647_S107_L002 0.00000000
37 ifc96.LIB018589_GEN00046648_S108_L002 0.00000000
38 ifc96.LIB018589_GEN00046649_S109_L002 0.00000000
39 ifc96.LIB018589_GEN00046650_S110_L002 0.00000000
40 ifc96.LIB018589_GEN00046651_S111_L002 0.00000000
data sample for annotation
sample_name Treatment Technology
1 bulk.DMSO-Pre-96IFC DMSO bulk
2 bulk.TSA-Pre-96IFC TSA bulk
3 bulk.DMSO-Pre-800IFC DMSO bulk
4 bulk.TSA-Pre-800IFC TSA bulk
5 bulk.DMSO-Post-800IFC DMSO bulk
6 bulk.TSA-Post-800IFC TSA bulk
7 ifc96.LIB018588_GEN00046618_S78_L001 TSA IFC96
8 ifc96.LIB018588_GEN00046619_S79_L001 TSA IFC96
9 ifc96.LIB018588_GEN00046620_S80_L001 TSA IFC96
10 ifc96.LIB018588_GEN00046621_S81_L001 TSA IFC96
11 ifc96.LIB018588_GEN00046622_S82_L001 TSA IFC96
12 ifc96.LIB018588_GEN00046623_S83_L001 TSA IFC96
13 ifc96.LIB018588_GEN00046624_S84_L001 TSA IFC96
14 ifc96.LIB018588_GEN00046625_S85_L001 TSA IFC96
15 ifc96.LIB018588_GEN00046626_S86_L001 TSA IFC96
16 ifc96.LIB018588_GEN00046627_S87_L001 TSA IFC96
17 ifc96.LIB018588_GEN00046628_S88_L001 TSA IFC96
18 ifc96.LIB018588_GEN00046629_S89_L001 TSA IFC96
19 ifc96.LIB018588_GEN00046630_S90_L001 TSA IFC96
20 ifc96.LIB018588_GEN00046631_S91_L001 TSA IFC96
21 ifc96.LIB018588_GEN00046632_S92_L001 TSA IFC96
22 ifc96.LIB018588_GEN00046633_S93_L001 TSA IFC96
23 ifc96.LIB018588_GEN00046634_S94_L001 TSA IFC96
24 ifc96.LIB018588_GEN00046635_S95_L001 TSA IFC96
25 ifc96.LIB018588_GEN00046636_S96_L001 TSA IFC96
26 ifc96.LIB018589_GEN00046637_S97_L002 DMSO IFC96
27 ifc96.LIB018589_GEN00046638_S98_L002 DMSO IFC96
28 ifc96.LIB018589_GEN00046639_S99_L002 DMSO IFC96
29 ifc96.LIB018589_GEN00046640_S100_L002 DMSO IFC96
30 ifc96.LIB018589_GEN00046641_S101_L002 DMSO IFC96
31 ifc96.LIB018589_GEN00046642_S102_L002 DMSO IFC96
32 ifc96.LIB018589_GEN00046643_S103_L002 DMSO IFC96
33 ifc96.LIB018589_GEN00046644_S104_L002 DMSO IFC96
34 ifc96.LIB018589_GEN00046645_S105_L002 DMSO IFC96
35 ifc96.LIB018589_GEN00046646_S106_L002 DMSO IFC96
36 ifc96.LIB018589_GEN00046647_S107_L002 DMSO IFC96
37 ifc96.LIB018589_GEN00046648_S108_L002 DMSO IFC96
38 ifc96.LIB018589_GEN00046649_S109_L002 DMSO IFC96
39 ifc96.LIB018589_GEN00046650_S110_L002 DMSO IFC96
40 ifc96.LIB018589_GEN00046651_S111_L002 DMSO IFC96
CodePudding user response:
First convert row names in the second dataframe to column called names, removing row names after it:
logtpm2$names <- rownames(logtpm2)
logtpm2 <- data.frame(logtpm2, row.names=NULL)
Then simply left_join the first dataframe with the second:
logtpm1 %>% left_join(logtpm2, by = "names")
Edit:
logtpm2 dataframe:
logtpm2 <- read.table(text = "sample_name Expression
bulk.DMSO.Pre.96IFC 0.05894781
bulk.TSA.Pre.96IFC 0
bulk.DMSO.Pre.800IFC 0
bulk.TSA.Pre.800IFC 0
bulk.DMSO.Post.800IFC 0
bulk.TSA.Post.800IFC 0
ifc96.LIB018588_GEN00046618_S78_L001 0
ifc96.LIB018588_GEN00046619_S79_L001 0
ifc96.LIB018588_GEN00046620_S80_L001 0
ifc96.LIB018588_GEN00046621_S81_L001 0
ifc96.LIB018588_GEN00046622_S82_L001 0
ifc96.LIB018588_GEN00046623_S83_L001 0
ifc96.LIB018588_GEN00046624_S84_L001 0
ifc96.LIB018588_GEN00046625_S85_L001 0
ifc96.LIB018588_GEN00046626_S86_L001 0
ifc96.LIB018588_GEN00046627_S87_L001 0
ifc96.LIB018588_GEN00046628_S88_L001 0
ifc96.LIB018588_GEN00046629_S89_L001 0
ifc96.LIB018588_GEN00046630_S90_L001 0
ifc96.LIB018588_GEN00046631_S91_L001 0
ifc96.LIB018588_GEN00046632_S92_L001 0
ifc96.LIB018588_GEN00046633_S93_L001 0
ifc96.LIB018588_GEN00046634_S94_L001 0
ifc96.LIB018588_GEN00046635_S95_L001 0
ifc96.LIB018588_GEN00046636_S96_L001 0
ifc96.LIB018589_GEN00046637_S97_L002 0
ifc96.LIB018589_GEN00046638_S98_L002 0
ifc96.LIB018589_GEN00046639_S99_L002 0
ifc96.LIB018589_GEN00046640_S100_L002 0
ifc96.LIB018589_GEN00046641_S101_L002 0
ifc96.LIB018589_GEN00046642_S102_L002 0
ifc96.LIB018589_GEN00046643_S103_L002 0
ifc96.LIB018589_GEN00046644_S104_L002 0
ifc96.LIB018589_GEN00046645_S105_L002 0
ifc96.LIB018589_GEN00046646_S106_L002 0
ifc96.LIB018589_GEN00046647_S107_L002 0
ifc96.LIB018589_GEN00046648_S108_L002 0
ifc96.LIB018589_GEN00046649_S109_L002 0
ifc96.LIB018589_GEN00046650_S110_L002 0
ifc96.LIB018589_GEN00046651_S111_L002 0
", header = TRUE)
logtpm1 dataframe:
logtpm1 <- read.table(text = "sample_name Treatment Technology
bulk.DMSO-Pre-96IFC DMSO bulk
bulk.TSA-Pre-96IFC TSA bulk
bulk.DMSO-Pre-800IFC DMSO bulk
bulk.TSA-Pre-800IFC TSA bulk
bulk.DMSO-Post-800IFC DMSO bulk
bulk.TSA-Post-800IFC TSA bulk
ifc96.LIB018588_GEN00046618_S78_L001 TSA IFC96
ifc96.LIB018588_GEN00046619_S79_L001 TSA IFC96
ifc96.LIB018588_GEN00046620_S80_L001 TSA IFC96
ifc96.LIB018588_GEN00046621_S81_L001 TSA IFC96
ifc96.LIB018588_GEN00046622_S82_L001 TSA IFC96
ifc96.LIB018588_GEN00046623_S83_L001 TSA IFC96
ifc96.LIB018588_GEN00046624_S84_L001 TSA IFC96
ifc96.LIB018588_GEN00046625_S85_L001 TSA IFC96
ifc96.LIB018588_GEN00046626_S86_L001 TSA IFC96
ifc96.LIB018588_GEN00046627_S87_L001 TSA IFC96
ifc96.LIB018588_GEN00046628_S88_L001 TSA IFC96
ifc96.LIB018588_GEN00046629_S89_L001 TSA IFC96
ifc96.LIB018588_GEN00046630_S90_L001 TSA IFC96
ifc96.LIB018588_GEN00046631_S91_L001 TSA IFC96
ifc96.LIB018588_GEN00046632_S92_L001 TSA IFC96
ifc96.LIB018588_GEN00046633_S93_L001 TSA IFC96
ifc96.LIB018588_GEN00046634_S94_L001 TSA IFC96
ifc96.LIB018588_GEN00046635_S95_L001 TSA IFC96
ifc96.LIB018588_GEN00046636_S96_L001 TSA IFC96
ifc96.LIB018589_GEN00046637_S97_L002 DMSO IFC96
ifc96.LIB018589_GEN00046638_S98_L002 DMSO IFC96
ifc96.LIB018589_GEN00046639_S99_L002 DMSO IFC96
ifc96.LIB018589_GEN00046640_S100_L002 DMSO IFC96
ifc96.LIB018589_GEN00046641_S101_L002 DMSO IFC96
ifc96.LIB018589_GEN00046642_S102_L002 DMSO IFC96
ifc96.LIB018589_GEN00046643_S103_L002 DMSO IFC96
ifc96.LIB018589_GEN00046644_S104_L002 DMSO IFC96
ifc96.LIB018589_GEN00046645_S105_L002 DMSO IFC96
ifc96.LIB018589_GEN00046646_S106_L002 DMSO IFC96
ifc96.LIB018589_GEN00046647_S107_L002 DMSO IFC96
ifc96.LIB018589_GEN00046648_S108_L002 DMSO IFC96
ifc96.LIB018589_GEN00046649_S109_L002 DMSO IFC96
ifc96.LIB018589_GEN00046650_S110_L002 DMSO IFC96
ifc96.LIB018589_GEN00046651_S111_L002 DMSO IFC96
", header = TRUE)
This command:
logtpm2 %>% left_join(logtpm1, by = "sample_name")
Returns this:
sample_name Expression Treatment Technology
1 bulk.DMSO.Pre.96IFC 0.05894781 <NA> <NA>
2 bulk.TSA.Pre.96IFC 0.00000000 <NA> <NA>
3 bulk.DMSO.Pre.800IFC 0.00000000 <NA> <NA>
4 bulk.TSA.Pre.800IFC 0.00000000 <NA> <NA>
5 bulk.DMSO.Post.800IFC 0.00000000 <NA> <NA>
6 bulk.TSA.Post.800IFC 0.00000000 <NA> <NA>
7 ifc96.LIB018588_GEN00046618_S78_L001 0.00000000 TSA IFC96
8 ifc96.LIB018588_GEN00046619_S79_L001 0.00000000 TSA IFC96
9 ifc96.LIB018588_GEN00046620_S80_L001 0.00000000 TSA IFC96
10 ifc96.LIB018588_GEN00046621_S81_L001 0.00000000 TSA IFC96
11 ifc96.LIB018588_GEN00046622_S82_L001 0.00000000 TSA IFC96
12 ifc96.LIB018588_GEN00046623_S83_L001 0.00000000 TSA IFC96
13 ifc96.LIB018588_GEN00046624_S84_L001 0.00000000 TSA IFC96
14 ifc96.LIB018588_GEN00046625_S85_L001 0.00000000 TSA IFC96
15 ifc96.LIB018588_GEN00046626_S86_L001 0.00000000 TSA IFC96
16 ifc96.LIB018588_GEN00046627_S87_L001 0.00000000 TSA IFC96
17 ifc96.LIB018588_GEN00046628_S88_L001 0.00000000 TSA IFC96
18 ifc96.LIB018588_GEN00046629_S89_L001 0.00000000 TSA IFC96
19 ifc96.LIB018588_GEN00046630_S90_L001 0.00000000 TSA IFC96
20 ifc96.LIB018588_GEN00046631_S91_L001 0.00000000 TSA IFC96
21 ifc96.LIB018588_GEN00046632_S92_L001 0.00000000 TSA IFC96
22 ifc96.LIB018588_GEN00046633_S93_L001 0.00000000 TSA IFC96
23 ifc96.LIB018588_GEN00046634_S94_L001 0.00000000 TSA IFC96
24 ifc96.LIB018588_GEN00046635_S95_L001 0.00000000 TSA IFC96
25 ifc96.LIB018588_GEN00046636_S96_L001 0.00000000 TSA IFC96
26 ifc96.LIB018589_GEN00046637_S97_L002 0.00000000 DMSO IFC96
27 ifc96.LIB018589_GEN00046638_S98_L002 0.00000000 DMSO IFC96
28 ifc96.LIB018589_GEN00046639_S99_L002 0.00000000 DMSO IFC96
29 ifc96.LIB018589_GEN00046640_S100_L002 0.00000000 DMSO IFC96
30 ifc96.LIB018589_GEN00046641_S101_L002 0.00000000 DMSO IFC96
31 ifc96.LIB018589_GEN00046642_S102_L002 0.00000000 DMSO IFC96
32 ifc96.LIB018589_GEN00046643_S103_L002 0.00000000 DMSO IFC96
33 ifc96.LIB018589_GEN00046644_S104_L002 0.00000000 DMSO IFC96
34 ifc96.LIB018589_GEN00046645_S105_L002 0.00000000 DMSO IFC96
35 ifc96.LIB018589_GEN00046646_S106_L002 0.00000000 DMSO IFC96
36 ifc96.LIB018589_GEN00046647_S107_L002 0.00000000 DMSO IFC96
37 ifc96.LIB018589_GEN00046648_S108_L002 0.00000000 DMSO IFC96
38 ifc96.LIB018589_GEN00046649_S109_L002 0.00000000 DMSO IFC96
39 ifc96.LIB018589_GEN00046650_S110_L002 0.00000000 DMSO IFC96
40 ifc96.LIB018589_GEN00046651_S111_L002 0.00000000 DMSO IFC96
Cases where Treatment or Technology is NA are the ones where there's no matching sample_name in logtpm1.
Edit 2:
As you have noticed, there are slight differences in sample_name spellings in the two datasets. Let's find which ones are not matching.
These sample_names are not found in logtpm2:
logtpm2$sample_name[!logtpm2$sample_name %in% logtpm1$sample_name]
[1] "bulk.DMSO.Pre.96IFC" "bulk.TSA.Pre.96IFC" "bulk.DMSO.Pre.800IFC" "bulk.TSA.Pre.800IFC" "bulk.DMSO.Post.800IFC" "bulk.TSA.Post.800IFC"
And these are not found in logtpm1:
logtpm1$sample_name[!logtpm1$sample_name %in% logtpm2$sample_name]
[1] "bulk.DMSO-Pre-96IFC" "bulk.TSA-Pre-96IFC" "bulk.DMSO-Pre-800IFC" "bulk.TSA-Pre-800IFC" "bulk.DMSO-Post-800IFC" "bulk.TSA-Post-800IFC"
Assuming you don't really care about dashes and dots, we can simply substitute all dashes with dots in sample_names of both datasets:
logtpm2$sample_name <- gsub('-', '.', logtpm2$sample_name)
logtpm1$sample_name <- gsub('-', '.', logtpm1$sample_name)
Now the same command:
library(dplyr)
logtpm2 %>% left_join(logtpm1, by = "sample_name")
will result in:
sample_name Expression Treatment Technology
1 bulk.DMSO.Pre.96IFC 0.05894781 DMSO bulk
2 bulk.TSA.Pre.96IFC 0.00000000 TSA bulk
3 bulk.DMSO.Pre.800IFC 0.00000000 DMSO bulk
4 bulk.TSA.Pre.800IFC 0.00000000 TSA bulk
5 bulk.DMSO.Post.800IFC 0.00000000 DMSO bulk
6 bulk.TSA.Post.800IFC 0.00000000 TSA bulk
7 ifc96.LIB018588_GEN00046618_S78_L001 0.00000000 TSA IFC96
8 ifc96.LIB018588_GEN00046619_S79_L001 0.00000000 TSA IFC96
9 ifc96.LIB018588_GEN00046620_S80_L001 0.00000000 TSA IFC96
10 ifc96.LIB018588_GEN00046621_S81_L001 0.00000000 TSA IFC96
11 ifc96.LIB018588_GEN00046622_S82_L001 0.00000000 TSA IFC96
12 ifc96.LIB018588_GEN00046623_S83_L001 0.00000000 TSA IFC96
13 ifc96.LIB018588_GEN00046624_S84_L001 0.00000000 TSA IFC96
14 ifc96.LIB018588_GEN00046625_S85_L001 0.00000000 TSA IFC96
15 ifc96.LIB018588_GEN00046626_S86_L001 0.00000000 TSA IFC96
16 ifc96.LIB018588_GEN00046627_S87_L001 0.00000000 TSA IFC96
17 ifc96.LIB018588_GEN00046628_S88_L001 0.00000000 TSA IFC96
18 ifc96.LIB018588_GEN00046629_S89_L001 0.00000000 TSA IFC96
19 ifc96.LIB018588_GEN00046630_S90_L001 0.00000000 TSA IFC96
20 ifc96.LIB018588_GEN00046631_S91_L001 0.00000000 TSA IFC96
21 ifc96.LIB018588_GEN00046632_S92_L001 0.00000000 TSA IFC96
22 ifc96.LIB018588_GEN00046633_S93_L001 0.00000000 TSA IFC96
23 ifc96.LIB018588_GEN00046634_S94_L001 0.00000000 TSA IFC96
24 ifc96.LIB018588_GEN00046635_S95_L001 0.00000000 TSA IFC96
25 ifc96.LIB018588_GEN00046636_S96_L001 0.00000000 TSA IFC96
26 ifc96.LIB018589_GEN00046637_S97_L002 0.00000000 DMSO IFC96
27 ifc96.LIB018589_GEN00046638_S98_L002 0.00000000 DMSO IFC96
28 ifc96.LIB018589_GEN00046639_S99_L002 0.00000000 DMSO IFC96
29 ifc96.LIB018589_GEN00046640_S100_L002 0.00000000 DMSO IFC96
30 ifc96.LIB018589_GEN00046641_S101_L002 0.00000000 DMSO IFC96
31 ifc96.LIB018589_GEN00046642_S102_L002 0.00000000 DMSO IFC96
32 ifc96.LIB018589_GEN00046643_S103_L002 0.00000000 DMSO IFC96
33 ifc96.LIB018589_GEN00046644_S104_L002 0.00000000 DMSO IFC96
34 ifc96.LIB018589_GEN00046645_S105_L002 0.00000000 DMSO IFC96
35 ifc96.LIB018589_GEN00046646_S106_L002 0.00000000 DMSO IFC96
36 ifc96.LIB018589_GEN00046647_S107_L002 0.00000000 DMSO IFC96
37 ifc96.LIB018589_GEN00046648_S108_L002 0.00000000 DMSO IFC96
38 ifc96.LIB018589_GEN00046649_S109_L002 0.00000000 DMSO IFC96
39 ifc96.LIB018589_GEN00046650_S110_L002 0.00000000 DMSO IFC96
40 ifc96.LIB018589_GEN00046651_S111_L002 0.00000000 DMSO IFC96
A full match as you can see. However, this is just a sample. Investigate all the names that are not matching in the two datasets (as I have shown before) and see what is the best way to perform substitutions of characters in sample_names of both dataframes to eliminate mismatching names.
CodePudding user response:
Is this what you are looking for?
library(dplyr)
library(stringr)
library(magrittr)
annotation %<>%
select(-Technology) %>%
rownames_to_column("Technology")
pattern <- paste0(unique(annotation$Treatment), collapse = "|")
logtpm2 %>%
left_join(annotation, by = "Technology") %>%
mutate(Treatment = ifelse(str_detect(Technology, pattern),
str_remove(str_sub(Technology, 6L, 9L),"\\."),
Treatment))
output:
Technology Expression Treatment
1 bulk.DMSO.Pre.96IFC 0.05894781 DMSO
2 bulk.TSA.Pre.96IFC 0.00000000 TSA
3 bulk.DMSO.Pre.800IFC 0.00000000 DMSO
4 bulk.TSA.Pre.800IFC 0.00000000 TSA
5 bulk.DMSO.Post.800IFC 0.00000000 DMSO
6 bulk.TSA.Post.800IFC 0.00000000 TSA
7 ifc96.LIB018588_GEN00046618_S78_L001 0.00000000 TSA
8 ifc96.LIB018588_GEN00046619_S79_L001 0.00000000 TSA
9 ifc96.LIB018588_GEN00046620_S80_L001 0.00000000 TSA
10 ifc96.LIB018588_GEN00046621_S81_L001 0.00000000 TSA
11 ifc96.LIB018588_GEN00046622_S82_L001 0.00000000 TSA
12 ifc96.LIB018588_GEN00046623_S83_L001 0.00000000 TSA
13 ifc96.LIB018588_GEN00046624_S84_L001 0.00000000 TSA
14 ifc96.LIB018588_GEN00046625_S85_L001 0.00000000 TSA
15 ifc96.LIB018588_GEN00046626_S86_L001 0.00000000 TSA
16 ifc96.LIB018588_GEN00046627_S87_L001 0.00000000 TSA
17 ifc96.LIB018588_GEN00046628_S88_L001 0.00000000 TSA
18 ifc96.LIB018588_GEN00046629_S89_L001 0.00000000 TSA
19 ifc96.LIB018588_GEN00046630_S90_L001 0.00000000 TSA
20 ifc96.LIB018588_GEN00046631_S91_L001 0.00000000 TSA
21 ifc96.LIB018588_GEN00046632_S92_L001 0.00000000 TSA
22 ifc96.LIB018588_GEN00046633_S93_L001 0.00000000 TSA
23 ifc96.LIB018588_GEN00046634_S94_L001 0.00000000 TSA
24 ifc96.LIB018588_GEN00046635_S95_L001 0.00000000 TSA
25 ifc96.LIB018588_GEN00046636_S96_L001 0.00000000 TSA
26 ifc96.LIB018589_GEN00046637_S97_L002 0.00000000 DMSO
27 ifc96.LIB018589_GEN00046638_S98_L002 0.00000000 DMSO
28 ifc96.LIB018589_GEN00046639_S99_L002 0.00000000 DMSO
29 ifc96.LIB018589_GEN00046640_S100_L002 0.00000000 DMSO
30 ifc96.LIB018589_GEN00046641_S101_L002 0.00000000 DMSO
31 ifc96.LIB018589_GEN00046642_S102_L002 0.00000000 DMSO
32 ifc96.LIB018589_GEN00046643_S103_L002 0.00000000 DMSO
33 ifc96.LIB018589_GEN00046644_S104_L002 0.00000000 DMSO
34 ifc96.LIB018589_GEN00046645_S105_L002 0.00000000 DMSO
35 ifc96.LIB018589_GEN00046646_S106_L002 0.00000000 DMSO
36 ifc96.LIB018589_GEN00046647_S107_L002 0.00000000 DMSO
37 ifc96.LIB018589_GEN00046648_S108_L002 0.00000000 DMSO
38 ifc96.LIB018589_GEN00046649_S109_L002 0.00000000 DMSO
39 ifc96.LIB018589_GEN00046650_S110_L002 0.00000000 DMSO
40 ifc96.LIB018589_GEN00046651_S111_L002 0.00000000 DMSO