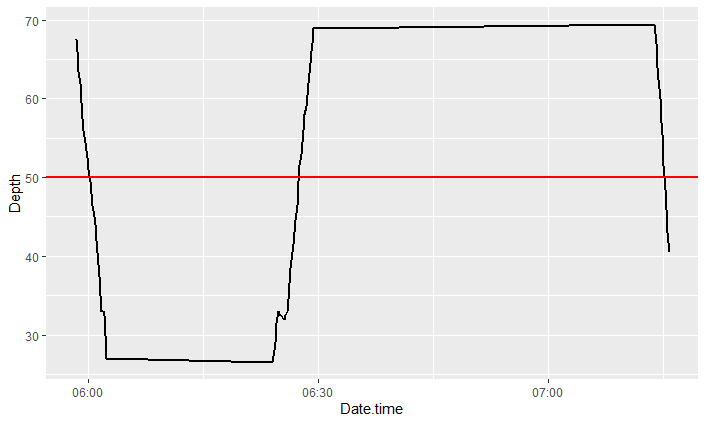
I am trying to calculate the amount of times my data (a line on a time series) crosses a certain threshold in R, preferably using dplyr. For this example I am trying to figure out how many times my data crosses a 50m threshold.
So in this case it would be 3 times that it crosses the 50m threshold. My dataframe is millions of points long so manually doing this is not an option. Any help would be much appreciated!
dput sample of data:
structure(list(Date.time = structure(c(1458626300, 1458626310,
1458626320, 1458626330, 1458626340, 1458626350, 1458626360, 1458626370,
1458626380, 1458626390, 1458626400, 1458626410, 1458626420, 1458626430,
1458626440, 1458626450, 1458626460, 1458626470, 1458626480, 1458626490,
1458626500, 1458626510, 1458626520, 1458626530, 1458626540, 1458627840,
1458627850, 1458627860, 1458627870, 1458627880, 1458627890, 1458627900,
1458627910, 1458627920, 1458627930, 1458627940, 1458627950, 1458627960,
1458627970, 1458627980, 1458627990, 1458628000, 1458628010, 1458628020,
1458628030, 1458628040, 1458628050, 1458628060, 1458628070, 1458628080,
1458628090, 1458628100, 1458628110, 1458628120, 1458628130, 1458628140,
1458628150, 1458628160, 1458630830, 1458630840, 1458630850, 1458630860,
1458630870, 1458630880, 1458630890, 1458630900, 1458630910, 1458630920,
1458630930, 1458630940), tzone = "UTC", class = c("POSIXct",
"POSIXt")), Depth = c(67.5, 66, 63.5, 62, 59.5, 57.5, 56, 54.5,
53.5, 52.5, 51, 49.5, 48, 46.5, 45, 44, 42, 40.5, 37.5, 35, 33,
33, 32.5, 30, 27, 26.5, 28, 29, 31.5, 33, 32.5, 32.5, 32.5, 32,
32, 32.5, 33, 34.5, 36.5, 38.5, 40.5, 41.5, 43, 44.5, 46.5, 49.5,
51.5, 53, 54.5, 56, 58, 59, 60.5, 62, 64.5, 66, 67, 69, 69.5,
67, 64, 62.5, 60, 57, 55.5, 51.5, 48.5, 46, 43, 40.5)), row.names = c(NA,
-70L), class = c("data.table", "data.frame")
CodePudding user response:
You can use data.table's rleid to identify which groups of data are below 50 and which are above. If you then take 1 away from the number of groups you get the amount of times the data passed the threshold:
library(data.table)
library(dplyr)
df %>%
summarize(rleid(Depth > 50)) %>%
n_distinct()-1
CodePudding user response:
This should do the trick too with dplyr:
You basicaly check for the curent value of Depth and the following one (using lead) are both side of the threshold and then count the number of time this condition is matched.
library(dplyr)
df <-
tibble(
Date.time =
structure(
c(
1458626300, 1458626310, 1458626320, 1458626330, 1458626340, 1458626350,
1458626360, 1458626370, 1458626380, 1458626390, 1458626400, 1458626410,
1458626420, 1458626430, 1458626440, 1458626450, 1458626460, 1458626470,
1458626480, 1458626490, 1458626500, 1458626510, 1458626520, 1458626530,
1458626540, 1458627840, 1458627850, 1458627860, 1458627870, 1458627880,
1458627890, 1458627900, 1458627910, 1458627920, 1458627930, 1458627940,
1458627950, 1458627960, 1458627970, 1458627980, 1458627990, 1458628000,
1458628010, 1458628020, 1458628030, 1458628040, 1458628050, 1458628060,
1458628070, 1458628080, 1458628090, 1458628100, 1458628110, 1458628120,
1458628130, 1458628140, 1458628150, 1458628160, 1458630830, 1458630840,
1458630850, 1458630860, 1458630870, 1458630880, 1458630890, 1458630900,
1458630910, 1458630920, 1458630930, 1458630940
),
tzone = "UTC", class = c("POSIXct", "POSIXt")
),
Depth =
c(
67.5, 66, 63.5, 62, 59.5, 57.5, 56, 54.5, 53.5, 52.5, 51, 49.5, 48,
46.5, 45, 44, 42, 40.5, 37.5, 35, 33, 33, 32.5, 30, 27, 26.5, 28, 29,
31.5, 33, 32.5, 32.5, 32.5, 32, 32, 32.5, 33, 34.5, 36.5, 38.5, 40.5,
41.5, 43, 44.5, 46.5, 49.5, 51.5, 53, 54.5, 56, 58, 59, 60.5, 62, 64.5,
66, 67, 69, 69.5, 67, 64, 62.5, 60, 57, 55.5, 51.5, 48.5, 46, 43, 40.5
)
)
## define the threshold
threshold <- 50
## keep only date just before threshold is crossed
df.cross <-
df %>%
filter(
## cross the treshold with positive trend
(Depth <= threshold & lead(Depth) > threshold) |
## or cross the treshold with negative trend
(Depth >= threshold & lead(Depth) < threshold)
)
df.cross
## count the number of time threshold is crossed
nrow(df.cross)
This should give you :
> df.cross
# A tibble: 3 × 2
Date.time Depth
<dttm> <dbl>
1 2016-03-22 06:00:00 51
2 2016-03-22 06:27:20 49.5
3 2016-03-22 07:15:00 51.5
> ## count the number of time threshold is crossed
> nrow(df.cross)
[1] 3
CodePudding user response:
Here's a dplyr way.
# dat1 is your data
thre <- 50
dat2 <- dat1 %>%
as_tibble() %>%
mutate(a1 = Depth > thre,
a2 = lag(Depth) < thre,
cross = a1 == a2)
sum(dat2$cross, na.rm = T)
CodePudding user response:
Tidyverse solution:
# Load the tidyverse package:
library(tidyverse)
# Set the threshold: th => integer scalar
th <- 50
# Calculate the number of times the threshold has been crossed:
# integer scalar => stdout(console)
df %>%
summarise(cnt = sum(
Depth >= th & lag(Depth) < th | Depth < th & lag(Depth) >= th,
na.rm = TRUE
)
) %>%
pull(cnt)
Base R solution:
# Set the threshold: th => integer scalar
th <- 50
# Calculate the number of times the threshold is crossed:
# integer scalar => stdout(console)
with(
df,
sum(Depth[-nrow(df)] >= th & Depth[-1] < th |
Depth[-nrow(df)] < th & Depth[-1] >= th)
)