I have a list of 14 lists of coefficients.. Each sublist is a quarter for a region. I need to plot the coefficients from 1-14 as X (progression of time) and the coefficients themselves as Y, with a different graph for each region. A reproducible example would be:
iris1 <- iris
iris2 <- iris
iris3 <- iris
iris_list <- list(iris, iris1, iris2, iris3)
res = lapply(iris_list, function(x){
test <- dlply(x, "Species", function(x) lm(Sepal.Length ~ Sepal.Width, data = x))
return(test)
})
Now, I need to plot [ Here, each graphic is a region(species), x is time. Thanks!
For clarification, i dont have to plot the intercept.
Here, each graphic is a region(species), x is time. Thanks!
For clarification, i dont have to plot the intercept.
CodePudding user response:
It is not very clear what you want to do, but I try to guess:
First, I changed your example to inject some randomness, so the line plot will not be a flat line. Second, I extracted from res the coefficient for each species for each sublist and transformed it into a long format dataframe (with reshape2::melt). Finally I plot the df using species as facetting variables
library(ggplot2)
library(plyr)
set.seed(4)
index <- lapply(1:4,function(x){sample(1:150,100,replace = T)})
iris_list <- lapply(index,function(ind){
iris[ind,]
})
res <- lapply(iris_list, function(x){
test <- plyr::dlply(x, "Species", function(x) lm(Sepal.Length ~ Sepal.Width, data = x))
return(test)
})
df <- as.data.frame(sapply(res,function(x){
do.call(rbind,lapply(x,coef))[,-1]
}))
df$species <- rownames(df)
m <- reshape2::melt(df)
m$variable <- as.integer(m$variable)
ggplot(m,aes(x=variable,y=value))
geom_line(aes(group=species))
facet_wrap(~species)
which give you this plot:
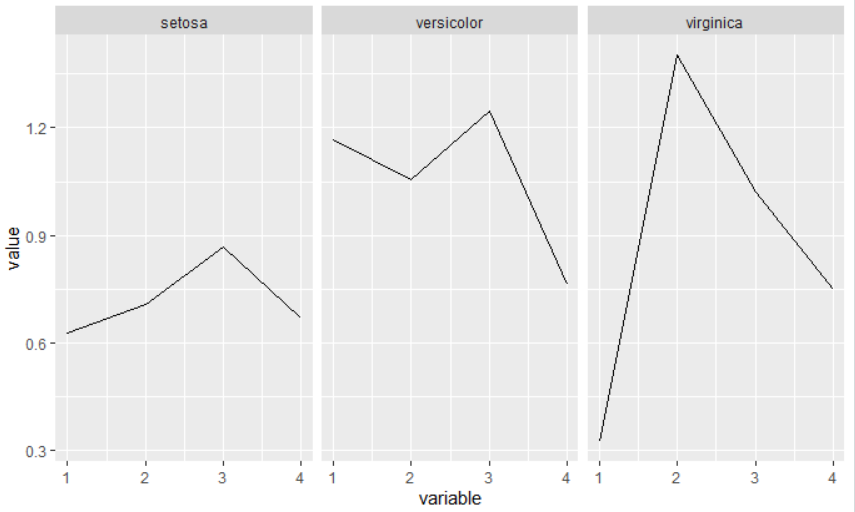 Now you have to customize the plot by changing
Now you have to customize the plot by changing theme options and adding col
