i have a dataframe where gene names are the column headers and sample names as rownames. they have one value for every cell. example:
G1 G2 G3
S1 0.19021657 0.129541639 0.741696359
S2 0.431098317 0.791633452 0.502163442
S3 0.433107132 0.085196075 0.071400934
S4 0.800781053 0.221191986 0.918588422
I would like to do a boxplot for every column (gene) - all plotted on one graph.
I tried to reshape the dataframe and then use ggplot, but I couldnt achieve the desired result. is there an elegant way to do this with ggplot?
CodePudding user response:
We can just use boxplot from base R on a data.frame. By default, it gives the boxplot for each columns of the dataset
boxplot(df1)
data
df1 <- structure(list(G1 = c(0.19021657, 0.431098317, 0.433107132, 0.800781053
), G2 = c(0.129541639, 0.791633452, 0.085196075, 0.221191986),
G3 = c(0.741696359, 0.502163442, 0.071400934, 0.918588422
)), class = "data.frame", row.names = c("S1", "S2", "S3",
"S4"))
CodePudding user response:
This type of problems generally has to do with reshaping the data. The format should be the long format and the data is in wide format. See 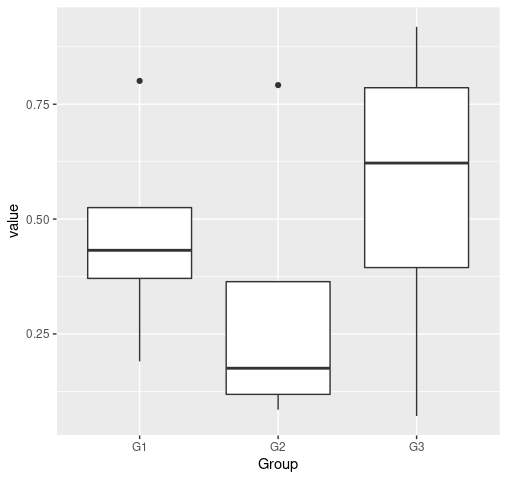
Data
df1 <- read.table(text = "
G1 G2 G3
S1 0.19021657 0.129541639 0.741696359
S2 0.431098317 0.791633452 0.502163442
S3 0.433107132 0.085196075 0.071400934
S4 0.800781053 0.221191986 0.918588422
", header = TRUE)
