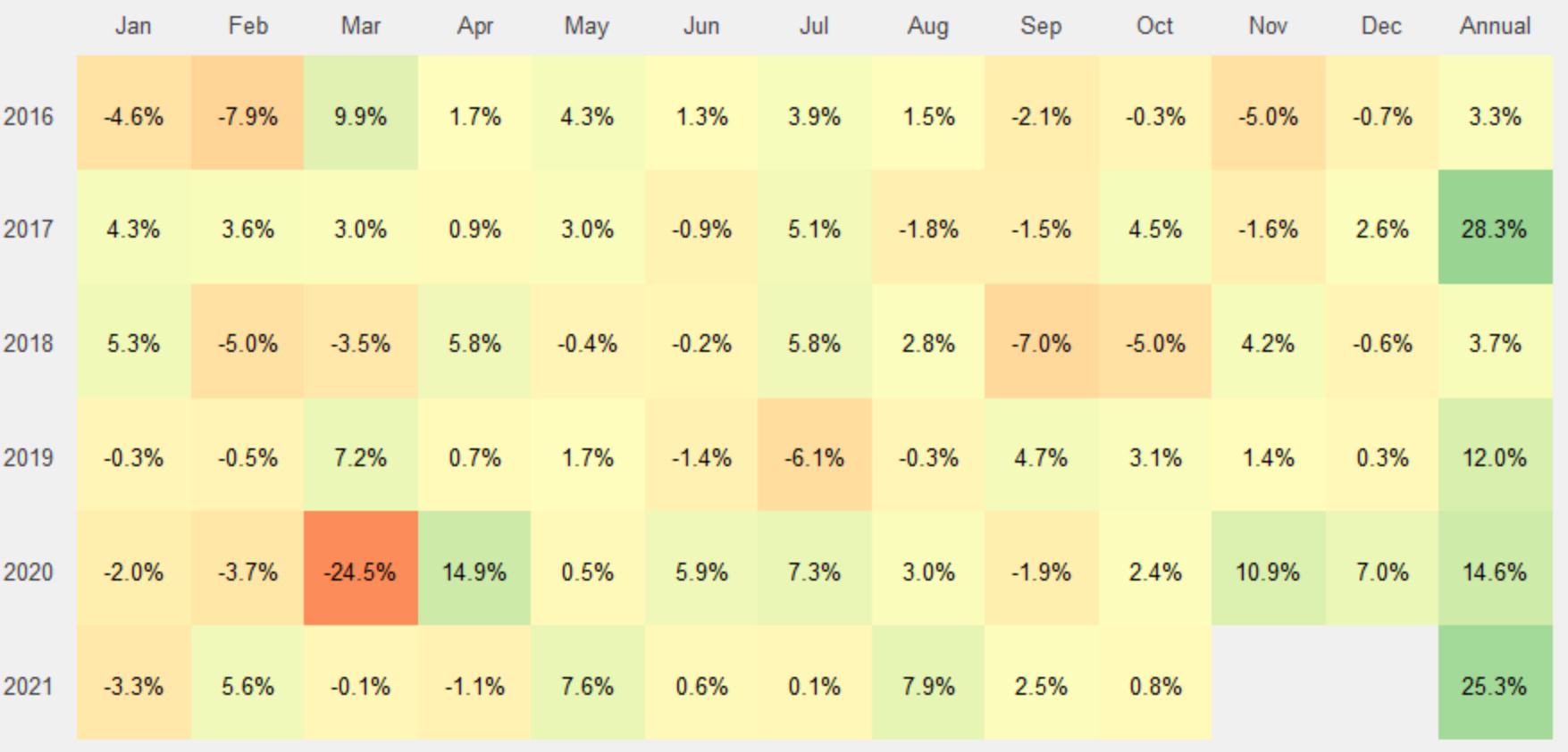
I have month column - Jan through Dec as well as 'Annual' value. I want to color heatmap based on Jan through Dec only (ignoring Annual). Values against Annual should be colored separately. In short gradient should be based on Jan through Dec.
Code below is working fine but it colors based on all unique values of month column.
Code
library(RColorBrewer)
library(ggplot2)
library(ggthemes)
myPalette <- colorRampPalette(brewer.pal(3, "Spectral"), space="Lab")
yaxis <- factor(df22$Year, levels = rev(unique(df22$Year)))
ggplot(df22, aes(x=Month, y=yaxis, fill=`Monthly Returns`)) geom_tile()
xlab("") ylab("")
guides(fill = FALSE)
scale_x_discrete(position = 'top')
scale_fill_gradientn(colours = myPalette(100))
theme_fivethirtyeight()
theme(
panel.grid.major = element_blank(),
panel.grid.minor = element_blank()
)
geom_text(label= scales::percent(df22$`Monthly Returns`,accuracy=0.1),
size = 3.5)
Data
structure(list(Month = structure(c(1L, 2L, 3L, 4L, 5L, 6L, 7L,
8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L,
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L,
4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L,
7L, 8L, 9L, 10L, 13L, 13L, 13L, 13L, 13L, 13L), .Label = c("Jan",
"Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct",
"Nov", "Dec", "Annual"), class = "factor"), Year = c(2016, 2016,
2016, 2016, 2016, 2016, 2016, 2016, 2016, 2016, 2016, 2016, 2017,
2017, 2017, 2017, 2017, 2017, 2017, 2017, 2017, 2017, 2017, 2017,
2018, 2018, 2018, 2018, 2018, 2018, 2018, 2018, 2018, 2018, 2018,
2018, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2020, 2020, 2020, 2020, 2020, 2020, 2020, 2020, 2020,
2020, 2020, 2020, 2021, 2021, 2021, 2021, 2021, 2021, 2021, 2021,
2021, 2021, 2016, 2017, 2018, 2019, 2020, 2021), `Monthly Returns` = c(-0.0455546382927932,
-0.0793794314513473, 0.0994778392356055, 0.0170703744247217,
0.0431308748372658, 0.0132714449349653, 0.039149313745751, 0.0152410238808452,
-0.0207479560262523, -0.0032483155414928, -0.0495369168026747,
-0.00705970342066962, 0.0427766059762552, 0.0360837088460483,
0.0302490450982238, 0.00905040881707368, 0.0301290066521883,
-0.00860613165737639, 0.0510171000111251, -0.0181317207157362,
-0.0149985938461858, 0.044676701273787, -0.0157646094851437,
0.0260140100477672, 0.0525075423394753, -0.0499522575153075,
-0.0349476851688416, 0.057891987545299, -0.0044232087547097,
-0.00224896419515394, 0.0581559876205109, 0.0282311484801734,
-0.0698913888620315, -0.0497946886834822, 0.0416646520083313,
-0.00623476893375712, -0.0034869756564318, -0.00542325250964097,
0.0720534160769843, 0.00711091062419609, 0.0168222388954323,
-0.0137948669664331, -0.0609718297586985, -0.00334082515221592,
0.0468481282064616, 0.031440487669275, 0.0142555652225132, 0.00258715177942692,
-0.0196728260436008, -0.0366116549940638, -0.244973563189387,
0.148623715952968, 0.00490898463313584, 0.0591404229656986, 0.0726138053972075,
0.0298393587023052, -0.0189065188181373, 0.0244578656451228,
0.108708436398415, 0.0703977730606203, -0.033305328712233, 0.0560050298648093,
-0.000802571331406262, -0.0113053287909243, 0.0760821911971872,
0.00587662594506211, 0.00050777370424187, 0.0792004231228312,
0.0251693738851843, 0.00797118377832791, 0.0329671724487319,
0.282651940477815, 0.0367452321549714, 0.119571125993303, 0.145843113875452,
0.252922033336702)), row.names = c(NA, -76L), class = c("tbl_df",
"tbl", "data.frame"))
CodePudding user response:
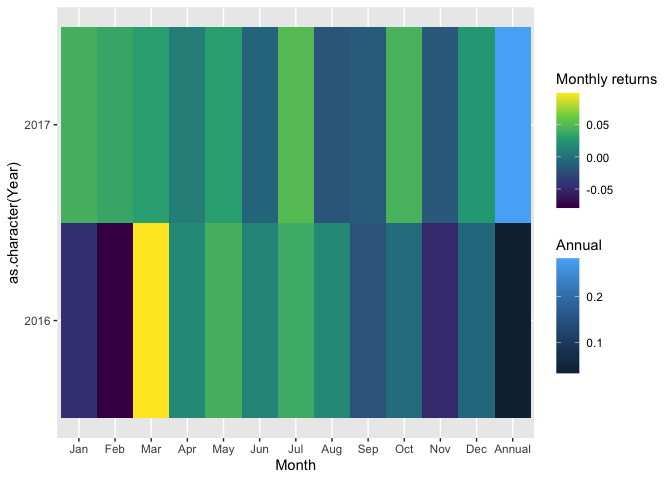
I've taken the liberty to simplify your data frame and also plotting code, trying to boil it down to the essential. I've renamed your returns column because it's much easier to code without back ticks, and instead I named the legend using the scale function.
What you're trying to achieve requires two separate fill scales for different parts of your data. Meaning, you need to plot the data separately, e.g. with two calls to geom_tile. You can create a second fill scale for example with ggnewscale.
In general, from both visualisation and coding perspective, I feel you might fare better by creating two plots separately, and stitch them together with for example patchwork. This will avoid the hassle of plotting different parts of your data, avoids the use of data frames that contain both values and aggregation and also will create a visual separation between measurements and aggregated values.
library(ggplot2)
ggplot(mapping = aes(x = Month, y = as.character(Year)))
geom_tile(data = subset(df22, Month != "Annual"), aes(fill = returns))
scale_fill_viridis_c("Monthly returns")
ggnewscale::new_scale_fill()
geom_tile(data = subset(df22, Month == "Annual"), aes(fill = returns))
scale_fill_continuous("Annual")
scale_x_discrete(limits = levels(df22$Month))
data
df22 <- structure(list(
Month = structure(c(
1L, 2L, 3L, 4L, 5L, 6L, 7L,
8L, 9L, 10L, 11L, 12L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L,
11L, 12L, 13L, 13L
), .Label = c(
"Jan", "Feb", "Mar", "Apr", "May",
"Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec", "Annual"
), class = "factor"),
Year = c(
2016, 2016, 2016, 2016, 2016, 2016, 2016, 2016,
2016, 2016, 2016, 2016, 2017, 2017, 2017, 2017, 2017, 2017,
2017, 2017, 2017, 2017, 2017, 2017, 2016, 2017
), returns = c(
-0.0455546382927932,
-0.0793794314513473, 0.0994778392356055, 0.0170703744247217,
0.0431308748372658, 0.0132714449349653, 0.039149313745751,
0.0152410238808452, -0.0207479560262523, -0.0032483155414928,
-0.0495369168026747, -0.00705970342066962, 0.0427766059762552,
0.0360837088460483, 0.0302490450982238, 0.00905040881707368,
0.0301290066521883, -0.00860613165737639, 0.0510171000111251,
-0.0181317207157362, -0.0149985938461858, 0.044676701273787,
-0.0157646094851437, 0.0260140100477672, 0.0329671724487319,
0.282651940477815
)
), row.names = c(NA, -26L), class = c(
"tbl_df",
"tbl", "data.frame"
))