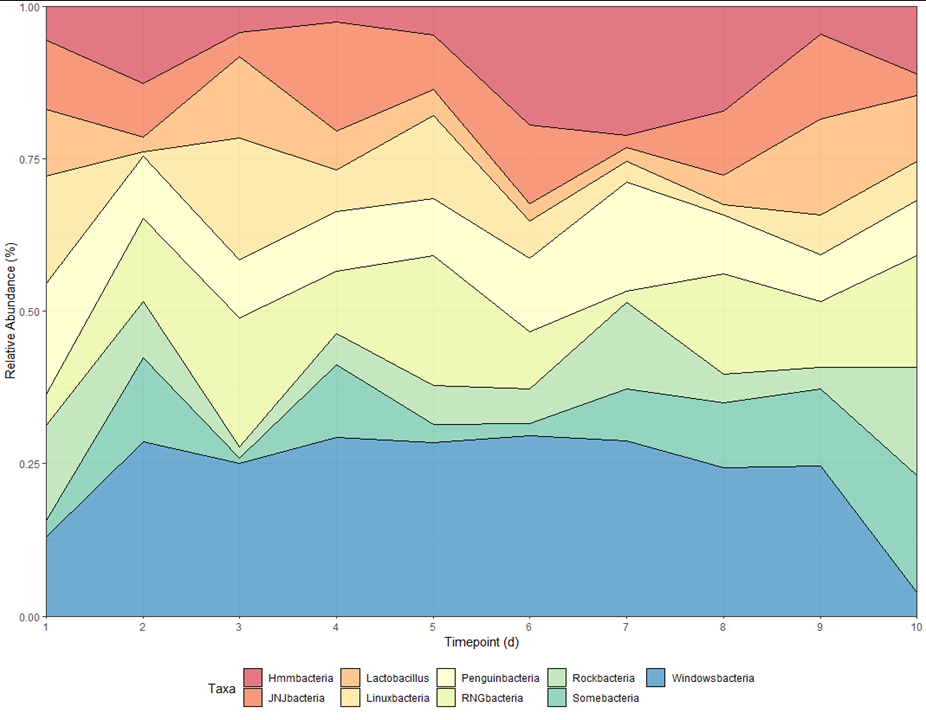
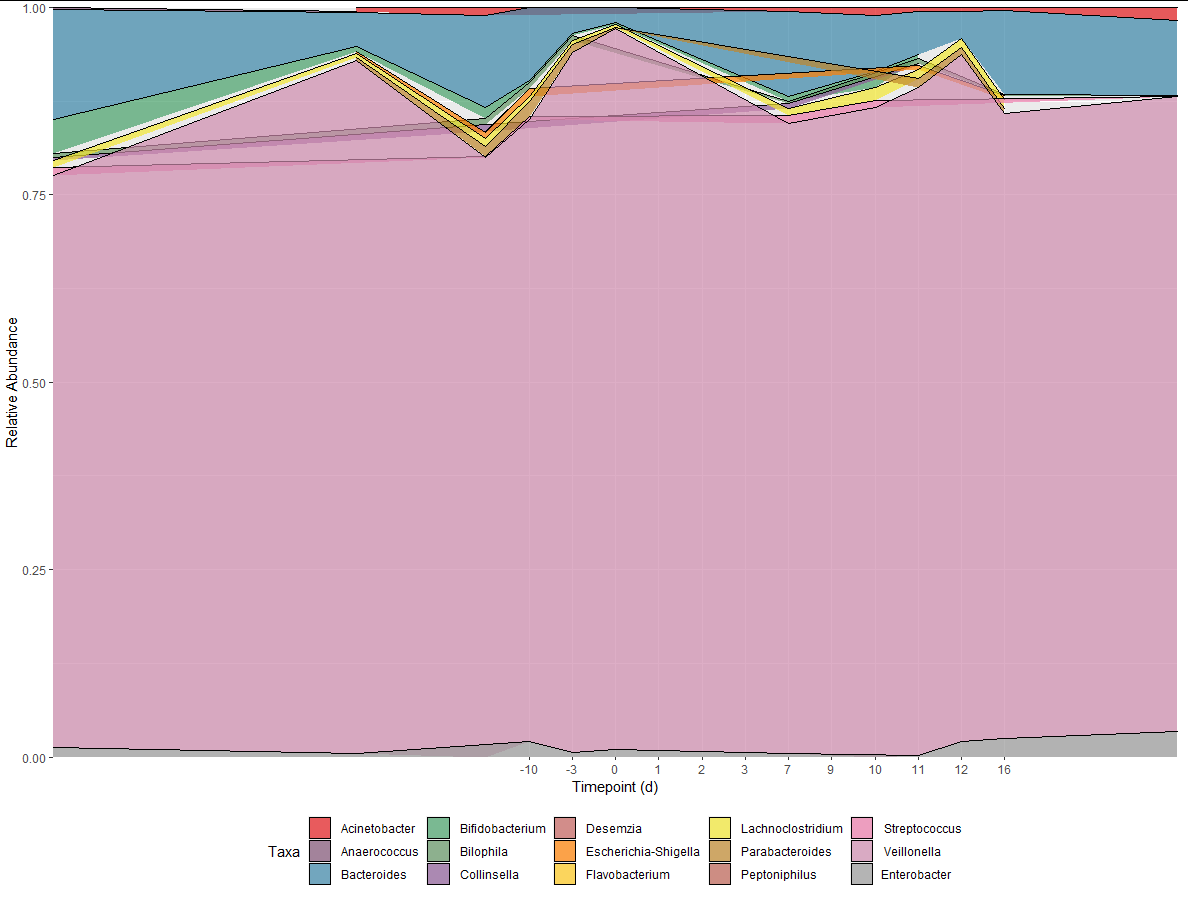
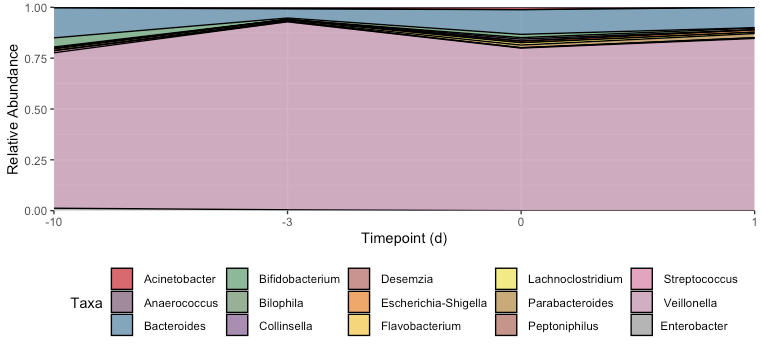
I'm trying to create a proportional stacked area graph as shown below in my mock data (Figure 1). When I try to do this with my real data, it comes out to Figure 2.
The class of the data are all the same after converting to percentages between the mock and 16S and are as follows: Timepoint - integer, Taxa - character, n - integer, percentage - numeric.
I'm looking to get the x-axis treated categorically and numerically (for two separate graphs) in the 16S data as with the mock and also to tidy up the overlapping lines (e.g., aesthetically the plot for 16S will look like the mock data).
dput(S1_RA1[1:40,])
structure(list(Timepoint = c(-10L, -10L, -10L, -10L, -10L, -10L,
-10L, -10L, -10L, -3L, -3L, -3L, -3L, -3L, -3L, -3L, -3L, -3L,
-3L, -3L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 0L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L), Taxa = c(" Anaerococcus", " Bacteroides",
" Bifidobacterium", " Bilophila", " Collinsella", " Lachnoclostridium",
" Streptococcus", " Veillonella", "Enterobacter", " Acinetobacter",
" Anaerococcus", " Bacteroides", " Bifidobacterium", " Escherichia-Shigella",
" Flavobacterium", " Lachnoclostridium", " Parabacteroides",
" Peptoniphilus", " Veillonella", "Enterobacter", " Acinetobacter",
" Bacteroides", " Bifidobacterium", " Bilophila", " Collinsella",
" Desemzia", " Escherichia-Shigella", " Lachnoclostridium", " Parabacteroides",
" Streptococcus", " Veillonella", " Bacteroides", " Bifidobacterium",
" Bilophila", " Desemzia", " Escherichia-Shigella", " Lachnoclostridium",
" Parabacteroides", " Streptococcus", " Veillonella"), n = c(40L,
2188L, 665L, 84L, 55L, 131L, 153L, 11325L, 185L, 127L, 62L, 1123L,
172L, 63L, 2L, 118L, 100L, 9L, 23123L, 109L, 253L, 2658L, 348L,
163L, 204L, 27L, 163L, 245L, 290L, 41L, 17497L, 2325L, 50L, 197L,
13L, 255L, 152L, 478L, 92L, 19692L), percentage = c(0.00269796303790638,
0.147578578173479, 0.0448536355051936, 0.0056657223796034, 0.00370969917712127,
0.0088358289491434, 0.0103197086199919, 0.763860785107244, 0.012478079050317,
0.00507837492002559, 0.00247920665387076, 0.0449056301983365,
0.00687779910428663, 0.00251919385796545, 7.99744081893794e-05,
0.00471849008317338, 0.00399872040946897, 0.000359884836852207,
0.92462412028151, 0.00435860524632118, 0.0115583169628581, 0.121430855680936,
0.0158983964548403, 0.0074466627072959, 0.00931974964594088,
0.00123349627666865, 0.0074466627072959, 0.0111928365845859,
0.0132486637123669, 0.00187308693864498, 0.799351272328567, 0.0978823727529154,
0.00210499726350356, 0.00829368921820402, 0.000547299288510925,
0.0107354860438681, 0.00639919168105081, 0.020123773839094, 0.00387319496484655,
0.829032122258241)), row.names = c(NA, -40L), groups = structure(list(
Timepoint = c(-10L, -3L, 0L, 1L), .rows = structure(list(
1:9, 10:20, 21:31, 32:40), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -4L), class = c("tbl_df",
"tbl", "data.frame"), .drop = TRUE), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"))
I've tried the following:
Setting the scale_x_discrete to scale_x_continuous
Converting aes(x = as.factor(Timepoint)..
Changing the limits/expand parameters in the scale_x_discrete code
Removing the negative timepoints
Changing the Number column in the S1_RA2 file to match the number system in Table 1
My code for the 16S is as follows and is almost identical to the mock except for the colors:
library(ggplot2)
library(dplyr)
RA1 <- read.csv("RA1.csv", header=TRUE)
#Transform relative abundance from RA1.csv to percentages
S1_RA1 <- RA1 %>%
group_by(Timepoint, Taxa) %>%
summarise(n = sum(Relative.Abundance)) %>%
mutate (percentage = n / sum(n))
head(Shime1_RA2)
#Set color palette to be able to include 15 colors
nb.cols <- 16
getPalette <- colorRampPalette(brewer.pal(9, 'Set1'))(nb.cols)
#Revised code - The code below works courtesy of Gregor's comment
library(tidyr)
Shime1_RA2 <- Shime1_RA2 %>% ungroup %>%
complete(Timepoint, Taxa, fill = list(n = 0, percentage = 0))
ggplot(Shime1_RA2, aes(x = factor(Timepoint), y = percentage, fill = Taxa, group = Taxa))
geom_area(position = "fill", colour = "black", size = .5, alpha = .7)
scale_y_continuous(name="Relative Abundance", expand=c(0,0))
scale_x_discrete(name="Timepoint (d)", expand=c(0,0))
scale_fill_manual(values = getPalette)
theme(legend.position='bottom')
CodePudding user response:
I fixed three things:
You want the x-scale to be treated categorically, so we need to
factor(Timepoint). (And then the default scale will be fine, so we delete your manually specifiedlimitsl)When we use a discrete x-axis scale, we have to explicitly tell
ggplotwhich dots we want to connect. We do this by adding thegroup = Taxaaesthetic.The weird lines cutting through the middle of other polygons are because you don't have an observation for every taxa at every timepoint, so when the dots are connected they may cut through intermediate timepoints. Use
tidyr::completeto fill in the missing observations with 0s.
library(tidyr)
S1_RA1 = S1_RA1 %>% ungroup %>%
complete(Timepoint, Taxa, fill = list(n = 0, percentage = 0))
ggplot(S1_RA1, aes(x = factor(Timepoint), y = percentage, fill = Taxa, group = Taxa))
geom_area(position = "fill", colour = "black", size = .5, alpha = .7)
scale_y_continuous(name="Relative Abundance", expand=c(0,0))
scale_x_discrete(
name="Timepoint (d)", expand=c(0,0)
)
scale_fill_manual(values = getPalette)
theme(legend.position='bottom')