I want to remove the for loop from the below code. In my use case, the value of n will be typically much larger where the value in the ranges of 200-700 is not uncommon, and it is inconvenient to list them all down, and adding one more loop would only make this more inefficient.
import numpy as np
n = 3
imgs = np.random.random((16,9,9,n))
transform = np.random.uniform(low=0.0, high=1.0, size=(n,n))
for img in imgs:
for channel in range(img.shape[2]):
temp1 = img[:,:,0]
temp2 = img[:,:,1]
temp3 = img[:,:,2]
temp = temp1 * transform[channel][0] temp2 * transform[channel][1] temp3 * transform[channel][2]
img[:,:,channel] = temp/3
Any pointers will be gratefully appreciated.
CodePudding user response:
I think you could maybe avoid the internal loop completely.
From my understanding you are taking a dot product of the transform matrix with the transpose of [temp1 temp2 temp3] matrix and then dividing it by 3.
The following is the representation of the same in an image:
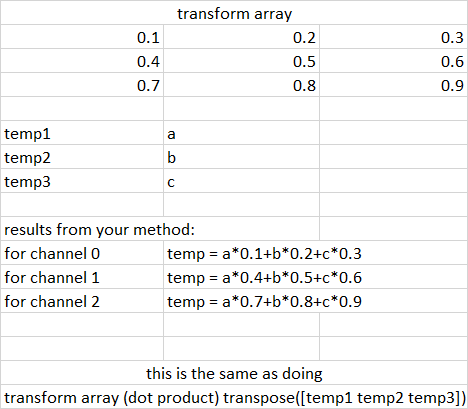
So, all of this could actually be done outside the for loop itself. The code for that would look something like this. P.S. Also edited the variable names at some places where they felt inconsistent.
import numpy as np
n = 3
imgs = np.random.random((16,9,9,n))
transform = np.random.uniform(low=0.0, high=1.0, size=(n,n))
for img in imgs:
temp_arr = img[:,:, 0:3]
img[:,:, 0:3] = np.dot(temp_arr, np.transpose(transform))/3
Compared the result against yours and it gives the same output
CodePudding user response:
Slicing might not be possible to be applied in the way you think it, but with numba you can get something that works without having to add another for loop.
This code performs the same operation as the code you provided.
import numpy as np
from numba import njit
@njit
def mult_arrays_values( array, values, n):
temp = np.zeros((array.shape[0], array.shape[1], n))
for i in range(n):
temp[:,:,i] = array[:,:,i] * values[i]
return temp
n = 3
imgs = np.random.random((16,9,9,n))
transform = np.random.uniform(low=0.0, high=1.0, size=(n,n))
for img in imgs:
for channel in range(img.shape[2]):
temp = mult_arrays_values(img[:,:,:], transform[channel][:], n)
temp = np.add.reduce(temp , axis=2)
img[:,:,channel] = temp/n
CodePudding user response:
You can also use numpy.einsum
In [77]: tr = np.arange(9).reshape((3,3))
...: imgs = np.arange(24).reshape((2,2,2,3)).transpose((1,0,2,3))
...: print('original imgs\n', imgs)
...: for img in imgs:
...: img[:,:,:] = np.dot(img, tr.T)
...: print('imgs after loop\n', imgs)
...: imgs = np.arange(24).reshape((2,2,2,3)).transpose((1,0,2,3))
...: print('result of einsum\n', np.einsum('ijkl,ml', imgs, tr))
original imgs
[[[[ 0 1 2]
[ 3 4 5]]
[[12 13 14]
[15 16 17]]]
[[[ 6 7 8]
[ 9 10 11]]
[[18 19 20]
[21 22 23]]]]
imgs after loop
[[[[ 5 14 23]
[ 14 50 86]]
[[ 41 158 275]
[ 50 194 338]]]
[[[ 23 86 149]
[ 32 122 212]]
[[ 59 230 401]
[ 68 266 464]]]]
result of einsum
[[[[ 5 14 23]
[ 14 50 86]]
[[ 41 158 275]
[ 50 194 338]]]
[[[ 23 86 149]
[ 32 122 212]]
[[ 59 230 401]
[ 68 266 464]]]]
