Dear All seniors and members,
Hope you are doing great. I have data set, which I like to plot the secondary x-axis in ggplot. I could not make it to work for the last 4 hours. below is my dataset.
Pathway ES NES p_value q_value Group
1 HALLMARK_HYPOXIA 0.49 2.25 0.000 0.000 Top
2 HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION 0.44 2.00 0.000 0.000 Top
3 HALLMARK_UV_RESPONSE_DN 0.45 1.98 0.000 0.000 Top
4 HALLMARK_TGF_BETA_SIGNALING 0.48 1.77 0.003 0.004 Top
5 HALLMARK_HEDGEHOG_SIGNALING 0.52 1.76 0.003 0.003 Top
6 HALLMARK_ESTROGEN_RESPONSE_EARLY 0.38 1.73 0.000 0.004 Top
7 HALLMARK_KRAS_SIGNALING_DN 0.37 1.69 0.000 0.005 Top
8 HALLMARK_INTERFERON_ALPHA_RESPONSE 0.37 1.54 0.009 0.021 Top
9 HALLMARK_TNFA_SIGNALING_VIA_NFKB 0.32 1.45 0.005 0.048 Top
10 HALLMARK_NOTCH_SIGNALING 0.42 1.42 0.070 0.059 Top
11 HALLMARK_COAGULATION 0.32 1.39 0.031 0.067 Top
12 HALLMARK_MITOTIC_SPINDLE 0.30 1.37 0.025 0.078 Top
13 HALLMARK_ANGIOGENESIS 0.40 1.37 0.088 0.074 Top
14 HALLMARK_WNT_BETA_CATENIN_SIGNALING 0.35 1.23 0.173 0.216 Top
15 HALLMARK_OXIDATIVE_PHOSPHORYLATION -0.65 -3.43 0.000 0.000 Bottom
16 HALLMARK_MYC_TARGETS_V1 -0.49 -2.56 0.000 0.000 Bottom
17 HALLMARK_E2F_TARGETS -0.45 -2.37 0.000 0.000 Bottom
18 HALLMARK_DNA_REPAIR -0.46 -2.33 0.000 0.000 Bottom
19 HALLMARK_ADIPOGENESIS -0.42 -2.26 0.000 0.000 Bottom
20 HALLMARK_FATTY_ACID_METABOLISM -0.41 -2.06 0.000 0.000 Bottom
21 HALLMARK_PEROXISOME -0.43 -2.01 0.000 0.000 Bottom
22 HALLMARK_MYC_TARGETS_V2 -0.43 -1.84 0.003 0.001 Bottom
23 HALLMARK_CHOLESTEROL_HOMEOSTASIS -0.42 -1.83 0.003 0.001 Bottom
24 HALLMARK_ALLOGRAFT_REJECTION -0.34 -1.78 0.000 0.003 Bottom
25 HALLMARK_MTORC1_SIGNALING -0.32 -1.67 0.000 0.004 Bottom
26 HALLMARK_P53_PATHWAY -0.29 -1.52 0.000 0.015 Bottom
27 HALLMARK_UV_RESPONSE_UP -0.28 -1.41 0.013 0.036 Bottom
28 HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY -0.35 -1.39 0.057 0.040 Bottom
29 HALLMARK_HEME_METABOLISM -0.26 -1.34 0.014 0.061 Bottom
30 HALLMARK_G2M_CHECKPOINT -0.23 -1.20 0.080 0.172 Bottom
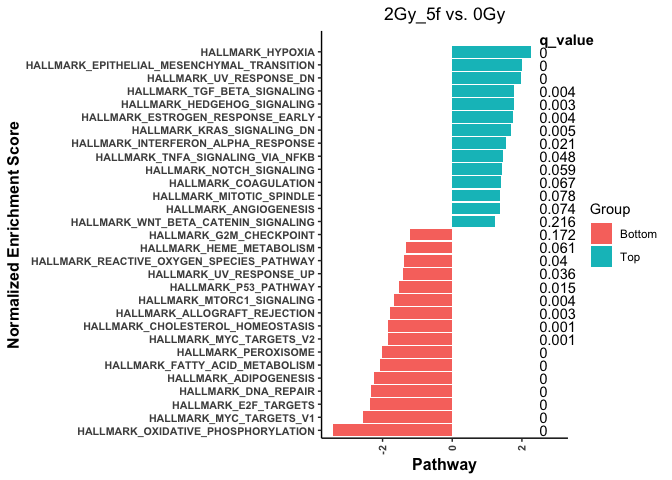
I like to plot like the following plot (plot # 1)
Here is my current codes chunks.
ggplot(data, aes(reorder(Pathway, NES), NES, fill= Group))
theme_classic() geom_col()
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1, size = 8),
axis.title = element_text(face = "bold", size = 12),
axis.text = element_text(face = "bold", size = 8), plot.title = element_text(hjust = 0.5)) labs(x="Pathway", y="Normalized Enrichment Score",
title="2Gy_5f vs. 0Gy") coord_flip()
This code produces the following plot (plot # 2)
So I would like to generate the plot where I have secondary x-axis with q_value (same like the first bar plot I have attached). Any help is greatly appreciated. Note: I used coord_flip so it turn angle of x-axis.
Kind Regards,
synat [1]: https://i.stack.imgur.com/dBFIS.jpg [2]: https://i.stack.imgur.com/yDbC5.jpg
CodePudding user response:
Maybe you don't need a secondary axis per se to get the plot style you seek.
library(tidyverse)
ggplot(data, aes(x = NES, y = reorder(Pathway, NES), fill= Group))
theme_classic()
geom_col()
geom_text(aes(x = 2.5, y = reorder(Pathway, NES), label = q_value), hjust = 0)
annotate("text", x = 2.5, y = length(data$Pathway) 1, hjust = 0, fontface = "bold", label = "q_value" )
coord_cartesian(xlim = c(NA, 3),
ylim = c(NA, length(data$Pathway) 1),
clip = "off")
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1, size = 8),
axis.title = element_text(face = "bold", size = 12),
axis.text = element_text(face = "bold", size = 8),
plot.title = element_text(hjust = 0.5))
labs(x="Pathway", y="Normalized Enrichment Score",
title="2Gy_5f vs. 0Gy")
And for future reference you can read in data in the format you pasted like so:
data <- read_table(
"
Pathway ES NES p_value q_value Group
HALLMARK_HYPOXIA 0.49 2.25 0.000 0.000 Top
HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION 0.44 2.00 0.000 0.000 Top
HALLMARK_UV_RESPONSE_DN 0.45 1.98 0.000 0.000 Top
HALLMARK_TGF_BETA_SIGNALING 0.48 1.77 0.003 0.004 Top
HALLMARK_HEDGEHOG_SIGNALING 0.52 1.76 0.003 0.003 Top
HALLMARK_ESTROGEN_RESPONSE_EARLY 0.38 1.73 0.000 0.004 Top
HALLMARK_KRAS_SIGNALING_DN 0.37 1.69 0.000 0.005 Top
HALLMARK_INTERFERON_ALPHA_RESPONSE 0.37 1.54 0.009 0.021 Top
HALLMARK_TNFA_SIGNALING_VIA_NFKB 0.32 1.45 0.005 0.048 Top
HALLMARK_NOTCH_SIGNALING 0.42 1.42 0.070 0.059 Top
HALLMARK_COAGULATION 0.32 1.39 0.031 0.067 Top
HALLMARK_MITOTIC_SPINDLE 0.30 1.37 0.025 0.078 Top
HALLMARK_ANGIOGENESIS 0.40 1.37 0.088 0.074 Top
HALLMARK_WNT_BETA_CATENIN_SIGNALING 0.35 1.23 0.173 0.216 Top
HALLMARK_OXIDATIVE_PHOSPHORYLATION -0.65 -3.43 0.000 0.000 Bottom
HALLMARK_MYC_TARGETS_V1 -0.49 -2.56 0.000 0.000 Bottom
HALLMARK_E2F_TARGETS -0.45 -2.37 0.000 0.000 Bottom
HALLMARK_DNA_REPAIR -0.46 -2.33 0.000 0.000 Bottom
HALLMARK_ADIPOGENESIS -0.42 -2.26 0.000 0.000 Bottom
HALLMARK_FATTY_ACID_METABOLISM -0.41 -2.06 0.000 0.000 Bottom
HALLMARK_PEROXISOME -0.43 -2.01 0.000 0.000 Bottom
HALLMARK_MYC_TARGETS_V2 -0.43 -1.84 0.003 0.001 Bottom
HALLMARK_CHOLESTEROL_HOMEOSTASIS -0.42 -1.83 0.003 0.001 Bottom
HALLMARK_ALLOGRAFT_REJECTION -0.34 -1.78 0.000 0.003 Bottom
HALLMARK_MTORC1_SIGNALING -0.32 -1.67 0.000 0.004 Bottom
HALLMARK_P53_PATHWAY -0.29 -1.52 0.000 0.015 Bottom
HALLMARK_UV_RESPONSE_UP -0.28 -1.41 0.013 0.036 Bottom
HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY -0.35 -1.39 0.057 0.040 Bottom
HALLMARK_HEME_METABOLISM -0.26 -1.34 0.014 0.061 Bottom
HALLMARK_G2M_CHECKPOINT -0.23 -1.20 0.080 0.172 Bottom")
Created on 2021-11-23 by the reprex package (v2.0.1)
