I'm am doing a programming assignment and have the following segments of code which should provide context to the data I'm using:
d <- read.csv(file='data6a.csv') # read and display data from CSV
d
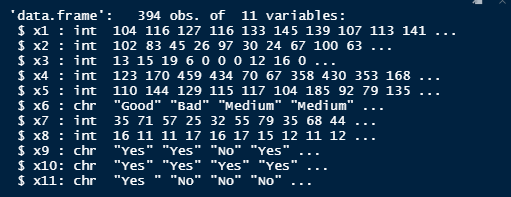
d = str(d)
d$colname=as.factor(d$colname)
The issue arises here. The assignment has given me this line of code to rename column x11 to y:
names(d)[names(d) == 'old.var.name'] <- 'new.var.name'
I have modified the code do to so:
names(d)[names(d) == 'x11'] <- 'y'
However, when I check to see that the column names are updated using str(d) as the assignment directs me, I am given the following output:
List of 1
$ colname: Factor w/ 0 levels:
This seems super straightforward yet I'm unsure why this is occurring. The code has basically been given to me other than that section I modified. If someone could point me in the right direction, that would be great.
CodePudding user response:
Things are getting messy from the point you're assigning d <-str(d) because str will return the summary of of the output but should not be assigned to an object, from that point d is not a dataframe anymore.
Just run the code as following
d <- read.csv(file='data6a.csv')
names(d)[names(d) == 'x11'] <- 'y'
If you want to view the data just run str(d) but do not assign it to anything
CodePudding user response:
Thanks to the help of @akrun, the issue was identified.
When performing d = str(d), d is set to null as str() does not return anything.
Changing the code to the following still has issues:
str(d)
d$colname=as.factor(d$colname)
as it returns:
Error in `$<-.data.frame`(`tmp`, colname, value = integer(0)) : >replacement has 0 rows, data has 394
Because this assignment requires me to redefine each categorical variable as a factor, we cannot skip over that segment and just change the column names using names(d)[names(d) == 'x11'] <- 'y.`
The solution that worked for me was the following:
temp = str(d)
temp$colname=as.factor(temp$colname)
The code above displayed the categorical columns as factors in the variable temp so we can apply the line names(d)[names(d) == 'x11'] <- 'y' to d as it originally was.