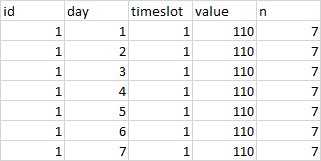
If the 'value' is equal (or the same) from day1 till day7 in dplyr how can I add up the n column? Also, how can I filter to have only n that are equal with 7
Preferable output:
Data sample:
structure(list(id = c(1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2,
2, 3, 3, 3, 3, 3, 3), day = c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3, 4,
5, 6, 7, 1, 2, 3, 4, 5, 6), timeslot = c(1, 1, 1, 1, 1, 1, 1,
2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3), value = c(110, 110, 110,
110, 110, 110, 110, 9990, 110, 110, 110, 110, 110, 9990, 110,
110, 110, 110, 8310, 110), n = c(1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L)), row.names = c(NA,
-20L), groups = structure(list(id = c(1, 1, 1, 1, 1, 1, 1, 2,
2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3), day = c(1, 2, 3, 4, 5, 6,
7, 1, 2, 3, 4, 5, 6, 7, 1, 2, 3, 4, 5, 6), timeslot = c(1, 1,
1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 3, 3, 3, 3, 3, 3), value = c(110,
110, 110, 110, 110, 110, 110, 9990, 110, 110, 110, 110, 110,
9990, 110, 110, 110, 110, 8310, 110), .rows = structure(list(
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L, 14L,
15L, 16L, 17L, 18L, 19L, 20L), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -20L), class = c("tbl_df",
"tbl", "data.frame"), .drop = TRUE), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"))
CodePudding user response:
Try this:
dat %>%
group_by(id) %>%
## test whether there is only one unique value per ID
filter(length(unique(value)) == 1) %>%
## sum up n
mutate(n = sum(n)) %>%
filter(n==7)
# A tibble: 7 × 5
# Groups: id [1]
id day timeslot value n
<dbl> <dbl> <dbl> <dbl> <int>
1 1 1 1 110 7
2 1 2 1 110 7
3 1 3 1 110 7
4 1 4 1 110 7
5 1 5 1 110 7
6 1 6 1 110 7
7 1 7 1 110 7
CodePudding user response:
library(data.table)
setDT(df)
df[, if (sum(n) == 7 && uniqueN(value) == 1) .SD, by = id]
#> id day timeslot value n
#> 1: 1 1 1 110 1
#> 2: 1 2 1 110 1
#> 3: 1 3 1 110 1
#> 4: 1 4 1 110 1
#> 5: 1 5 1 110 1
#> 6: 1 6 1 110 1
#> 7: 1 7 1 110 1
Created on 2021-11-23 by the reprex package (v2.0.1)
CodePudding user response:
I'll sneak in a little data.table approach because that's always popular - it is counting the number of observations and number of unique values (per id for both of those), then generating a data.table with id and the logical conditions evaluated, which is then merged onto the original data, which is finally filtered.
library(data.table)
setDT(data1)
data1[data1[, .(.N, uniqueN(value)), by=id][, .(id, N==7 & V2==1)], on="id"][V2==TRUE, -c("V2")]
Edit: Credit to IceCreamToucan for the main work in this solution - I just wanted to add a bit of detail around it and make some slight tweaks. This puts an if statement into the j of DT[i, j, by] (see[(https://cran.r-project.org/web/packages/data.table/vignettes/datatable-intro.html) for details). When that if statement resolves to TRUE, it is returning the selected variables given by the special character, .SD — .SDcols was missing so it defaulted to all columns. This version also uses the .N special character — a synonym for nrows(). This j-process is done by id.
data1[, if(.N==7 & uniqueN(value)==1){.SD}, by=id]
CodePudding user response:
Let's name your dataframe as df.
So you can use this:
# Add up column "n"
df %>%
left_join(df %>% group_by(id) %>% summarise(total_n = sum(n)) ) %>%
select(everything(), - n) %>%
filter(total_n == 7)
Output:
# A tibble: 20 x 5
# Groups: id, day, timeslot, value [20]
id day timeslot value total_n
<dbl> <dbl> <dbl> <dbl> <int>
1 1 1 1 110 7
2 1 2 1 110 7
3 1 3 1 110 7
4 1 4 1 110 7
5 1 5 1 110 7
6 1 6 1 110 7
7 1 7 1 110 7
8 2 1 2 9990 7
9 2 2 2 110 7
10 2 3 2 110 7
11 2 4 2 110 7
12 2 5 2 110 7
13 2 6 2 110 7
14 2 7 2 9990 7
CodePudding user response:
Update after clarification (thanks to rg255):
library(dplyr)
df %>%
group_by(id) %>%
mutate(n=ifelse(n_distinct(value)==1, n(), 0)) %>%
filter(n == 7)
id day timeslot value n
<dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 1 110 7
2 1 2 1 110 7
3 1 3 1 110 7
4 1 4 1 110 7
5 1 5 1 110 7
6 1 6 1 110 7
7 1 7 1 110 7