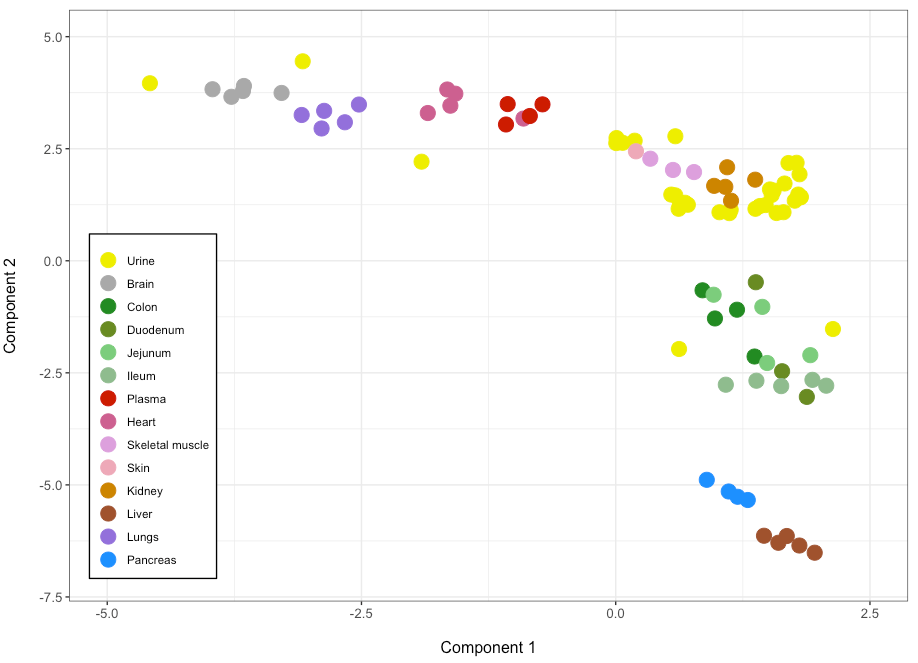
I have the following table:
Sample UMAP1 UMAP2 colors tissue
C_A1 1.41771557752425 1.22200433730937 yellow2 Urine
C_A2 1.76055361418532 1.34108703383903 yellow2 Urine
C_A3 0.00750224109526362 2.73864657429924 yellow2 Urine
C_A4 1.6500702891979 1.08443364179582 yellow2 Urine
C_A5 1.47433412636331 1.24338667538958 yellow2 Urine
C_A9 1.81676319795816 1.41560757063772 yellow2 Urine
C_B1 1.58085792476313 1.06578750509132 yellow2 Urine
C_B2 0.54619869458667 1.47703917280933 yellow2 Urine
CKD_C1 1.77876535894167 2.1866355905902 yellow2 Urine
CKD_C2 1.5528582393691 1.56720266537748 yellow2 Urine
CKD_C3 0.00527727382088514 2.62619847720534 yellow2 Urine
CKD_C4 0.586308172828281 2.7784840588712 yellow2 Urine
CKD_C5 0.186228467054963 2.6796009437192 yellow2 Urine
PYL_E1 -3.07772498762527 4.45095110253223 yellow2 Urine
PYL_E2 1.1175301960485 1.06349707623584 yellow2 Urine
PYL_E3 1.82042047858787 1.42547383706531 yellow2 Urine
PYL_E4 1.66009595777039 1.72432475907437 yellow2 Urine
PYL_E5 1.13056114256336 1.13760951795915 yellow2 Urine
PYL_E6 1.79463825137017 1.47814506344302 yellow2 Urine
PYL_E7 0.622690635767324 -1.96794491349824 yellow2 Urine
PYL_E8 0.63118627267307 1.29871116070517 yellow2 Urine
PYL_E9 -4.58033059073059 3.96133161881786 yellow2 Urine
PYL_F1 1.51666575038286 1.59237710006909 yellow2 Urine
UO_D1 0.709257524588014 1.24974065107936 yellow2 Urine
UO_D2 1.69833621648934 2.18100899996296 yellow2 Urine
UO_D3 1.01987315310411 1.08349161107457 yellow2 Urine
UO_D4 1.53357549894633 1.46876538726954 yellow2 Urine
UO_D5 0.618372187901223 1.1600432701727 yellow2 Urine
UO_D6 1.80740300575174 1.93028754079873 yellow2 Urine
UO_D7 -1.90935774520858 2.21114064992169 yellow2 Urine
SBU_G1 0.0684949931212032 2.6323213276562 yellow2 Urine
SBU_G2 1.37378744895387 1.15990665575179 yellow2 Urine
SBU_G3 0.681350048991714 1.29504193314325 yellow2 Urine
SBU_G4 0.585511550079965 1.46021542902406 yellow2 Urine
SBU_G5 2.13609698314454 -1.52225184779846 yellow2 Urine
BR1.RPM -3.28586643197682 3.74476178876053 darkgrey Brain
BR2.RPM -3.66581644761413 3.78800081614875 darkgrey Brain
BR3.RPM -3.96475462680686 3.8275429988704 darkgrey Brain
BR4.RPM -3.6558646156002 3.90023979275899 darkgrey Brain
BR5.RPM -3.77917418926683 3.65892915155001 darkgrey Brain
CO1.RPM 1.19249345974165 -1.0914160958934 forestgreen Colon
CO2.RPM 0.853658643619059 -0.658496441780366 forestgreen Colon
CO3.RPM 0.975760223303845 -1.28665248361392 forestgreen Colon
CO5.RPM 1.36511790751794 -2.13567084452923 forestgreen Colon
DU1.RPM 1.63592949492624 -2.46405219183458 olivedrab4 Duodenum
DU2.RPM 1.87925711931471 -3.03633498025664 olivedrab4 Duodenum
DU5.RPM 1.37754729522807 -0.478401566279114 olivedrab4 Duodenum
HE1.RPM -1.84787050174421 3.29683025222076 hotpink3 Heart
HE2.RPM -1.57712374669721 3.72779965312866 hotpink3 Heart
HE3.RPM -1.62595902698841 3.4606008066362 hotpink3 Heart
HE4.RPM -0.907119493708492 3.17316852377154 hotpink3 Heart
HE5.RPM -1.65644593797311 3.82195960352629 hotpink3 Heart
IL1.RPM 1.44153778801558 -1.02823513385223 palegreen3 Ileum
IL2.RPM 1.4889578583487 -2.2804483432248 palegreen3 Ileum
IL3.RPM 1.91342848325147 -2.10569098957249 palegreen3 Ileum
IL4.RPM 0.962529810173936 -0.757115204974117 palegreen3 Ileum
JE1.RPM 1.93437797225407 -2.65674160788117 darkseagreen Jejunum
JE2.RPM 2.06996643344844 -2.78640061968243 darkseagreen Jejunum
JE3.RPM 1.62706829576533 -2.79624294732553 darkseagreen Jejunum
JE4.RPM 1.0825326758516 -2.76470251848877 darkseagreen Jejunum
JE5.RPM 1.38342677881686 -2.6755546386966 darkseagreen Jejunum
KI1.RPM 0.967578089848615 1.67184647007406 orange3 Kidney
KI2.RPM 1.13378641637209 1.34124080803019 orange3 Kidney
KI3.RPM 1.37130716354727 1.80996835872441 orange3 Kidney
KI4.RPM 1.07772033561742 1.64994468114772 orange3 Kidney
KI5.RPM 1.0955496159798 2.0869739595772 orange3 Kidney
LI1.RPM 1.682084870402 -6.14052679905332 sienna Liver
LI2.RPM 1.59906448215381 -6.29131805082633 sienna Liver
LI3.RPM 1.45802709716468 -6.13448078364028 sienna Liver
LI4.RPM 1.95665039443957 -6.5130493984287 sienna Liver
LI5.RPM 1.80607410919614 -6.35273035030961 sienna Liver
LU1.RPM -2.52491770797571 3.4865019812581 mediumpurple Lung
LU2.RPM -2.86699310757743 3.34528752430189 mediumpurple Lung
LU3.RPM -2.89232280631143 2.95189454920787 mediumpurple Lung
LU4.RPM -2.66332768628549 3.09050672631981 mediumpurple Lung
LU5.RPM -3.08812503849215 3.25327915431485 mediumpurple Lung
PA1.RPM 1.29849330321564 -5.33710072865804 dodgerblue Pancreas
PA2.RPM 1.19873085339097 -5.26535178376946 dodgerblue Pancreas
PA3.RPM 1.11108191265227 -5.14709232997355 dodgerblue Pancreas
PA5.RPM 0.895423334114921 -4.88663713272289 dodgerblue Pancreas
PL1.RPM -1.06236761107225 3.49621079235936 red3 Blood
PL2.RPM -1.07888887772428 3.04101229356896 red3 Blood
PL3.RPM -0.844686506146655 3.23004986615368 red3 Blood
PL5.RPM -0.720030063721917 3.49005127885431 red3 Blood
SKI.RPM 0.199109542570694 2.44218482056362 pink2 Skin
SM1.RPM 0.563134748949276 2.02657901642232 plum Muscle
SM2.RPM 0.340223402531866 2.27754829463276 plum Muscle
SM3.RPM 0.770024511959072 1.97953000238431 plum Muscle
And the following ggplot2 code:
ggplot(Cat_urine, aes(x=UMAP1, y=UMAP2, color=colors))
geom_point(size=5)
scale_color_manual(labels = c("Urine","Brain", "Colon", "Duodenum", "Jejunum", "Ileum", "Plasma", "Heart", "Skeletal muscle", "Skin",
"Kidney", "Liver", "Lungs", "Pancreas", "Urine"),
values = c("yellow2","darkgrey", "forestgreen", "olivedrab4", "palegreen3", "darkseagreen", "red3", "hotpink3", "plum", "pink2",
"orange3", "sienna", "mediumpurple", "dodgerblue"))
guides(colour = guide_legend(override.aes = list(size=5)))
ylim(c(-7,5)) xlim(c(-5,2.5)) guides(shape = guide_legend(override.aes = list(size = 4)))
theme_bw() theme(axis.text.x = element_text(size=10), axis.title=element_text(size=12),
axis.text.y =element_text(size=10),plot.margin = margin(1, 0.5, 0.5, 1, "cm"))
xlab("\nComponent 1") ylab("Component 2\n")
theme(legend.position = c(0.1, 0.33), legend.title = element_blank(),
legend.background = element_rect(fill = "white", color = "black"))
Which generates this figure:
I would like to divide the legend in two columns of 7 elements each. I have tried with guides(fill=guide_legend(ncol=2)) but it did not modify anything. Maybe I put it in the wrong place or it might be another way to achieve that. Could anyone help?
Many thanks
CodePudding user response:
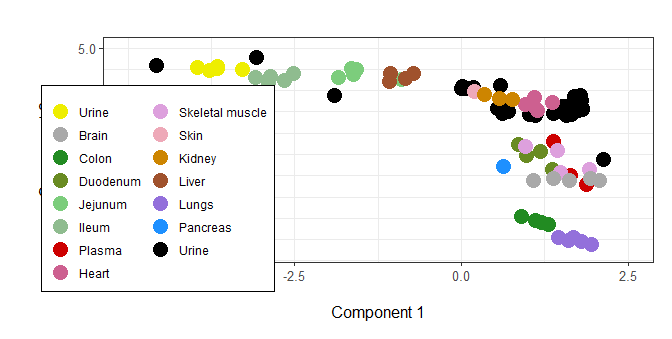
You never define fill= as an aesthetic; use guides(color=...) instead.
Note: with this sample data, I needed to add another color to the scale_color_manual; it shouldn't be necessary with your real data. The only change I'm adding to your code is one argument to guides.
ggplot(Cat_urine, aes(x=UMAP1, y=UMAP2, color=colors))
geom_point(size=5)
scale_color_manual(labels = c("Urine","Brain", "Colon", "Duodenum", "Jejunum", "Ileum", "Plasma", "Heart", "Skeletal muscle", "Skin",
"Kidney", "Liver", "Lungs", "Pancreas", "Urine"),
values = c("yellow2","darkgrey", "forestgreen", "olivedrab4", "palegreen3", "darkseagreen", "red3", "hotpink3", "plum", "pink2",
"orange3", "sienna", "mediumpurple", "dodgerblue", "black"))
guides(
colour = guide_legend(ncol = 2, override.aes = list(size=5))
## ^^^^^^ ^^^^^^^^ these are the changes
)
ylim(c(-7,5)) xlim(c(-5,2.5)) guides(shape = guide_legend(override.aes = list(size = 4)))
theme_bw()
theme(axis.text.x = element_text(size=10), axis.title=element_text(size=12),
axis.text.y =element_text(size=10),plot.margin = margin(1, 0.5, 0.5, 1, "cm"))
xlab("\nComponent 1")
ylab("Component 2\n")
theme(legend.position = c(0.1, 0.33), legend.title = element_blank(),
legend.background = element_rect(fill = "white", color = "black"))
Granted, you'll need to adjust the legend location ...