How to replace the default geom_ribbon with geom_errorbar in ggcompetingrisks from survminer package?
conf.int = T will put confidence interval as a ribbon layer.
my code:
library(cmprsk);library(survminer)
set.seed(2)
ss <- rexp(100)
gg <- factor(sample(1:3,100,replace=TRUE),1:3,c('BRCA','LUNG','OV'))
cc <- factor(sample(0:2,100,replace=TRUE),0:2,c('no event', 'death', 'progression'))
strt <- sample(1:2,100,replace=TRUE)
# handles cuminc objects
print(fit <- cmprsk::cuminc(ss,cc,gg,strt))
ggcompetingrisks(fit, multiple_panels = FALSE, conf.int = TRUE)
Any advice will be greatly appreciated.
CodePudding user response:
You can modify the source code of the two required functions from the survminer package (ggcompetingrisks.cuminc() & ggcompetingrisks()), e.g.
#install.packages("cmprsk")
library(cmprsk)
#> Loading required package: survival
library(survminer)
#> Loading required package: ggplot2
#> Loading required package: ggpubr
#>
#> Attaching package: 'survminer'
#> The following object is masked from 'package:survival':
#>
#> myeloma
set.seed(2)
ss <- rexp(100)
gg <- factor(sample(1:3,100,replace=TRUE),1:3,c('BRCA','LUNG','OV'))
cc <- factor(sample(0:2,100,replace=TRUE),0:2,c('no event', 'death', 'progression'))
strt <- sample(1:2,100,replace=TRUE)
# handles cuminc objects
print(fit <- cmprsk::cuminc(ss,cc,gg,strt))
#> Tests:
#> stat pv df
#> no event 0.008709109 0.9956549 2
#> death 1.629098402 0.4428389 2
#> progression 2.605007849 0.2718502 2
#> Estimates and Variances:
#> $est
#> 1 2 3 4
#> BRCA no event 0.17241379 0.2758621 0.2758621 0.2758621
#> LUNG no event 0.25641026 0.3076923 0.3076923 0.3333333
#> OV no event 0.15625000 0.2500000 0.3125000 0.3125000
#> BRCA death 0.24137931 0.2758621 0.2758621 0.2758621
#> LUNG death 0.07692308 0.2307692 0.2307692 0.2307692
#> OV death 0.21875000 0.3437500 0.3750000 0.3750000
#> BRCA progression 0.34482759 0.4137931 0.4137931 0.4137931
#> LUNG progression 0.25641026 0.3589744 0.3846154 0.3846154
#> OV progression 0.18750000 0.2812500 0.2812500 0.2812500
#>
#> $var
#> 1 2 3 4
#> BRCA no event 0.005277565 0.007933520 0.007933520 0.007933520
#> LUNG no event 0.005083701 0.005740953 0.005740953 0.006209164
#> OV no event 0.004338366 0.006323011 0.007854783 0.007854783
#> BRCA death 0.006783761 0.007478906 0.007478906 0.007478906
#> LUNG death 0.001887310 0.004961788 0.004961788 0.004961788
#> OV death 0.005593145 0.007641923 0.008089627 0.008089627
#> BRCA progression 0.008299977 0.009699862 0.009699862 0.009699862
#> LUNG progression 0.005097395 0.006324651 0.006608777 0.006608777
#> OV progression 0.004972034 0.006833076 0.006833076 0.006833076
ggcompetingrisks(fit, multiple_panels = FALSE, conf.int = TRUE)
ggcompetingrisks_cuminc_altered <- function(fit, gnames = NULL, gsep = " ", multiple_panels = TRUE,
coef = 1.96, conf.int = FALSE)
{
if (!is.null(fit$Tests))
fit <- fit[names(fit) != "Tests"]
fit2 <- lapply(fit, `[`, 1:3)
if (is.null(gnames))
gnames <- names(fit2)
fit2_list <- lapply(seq_along(gnames), function(ind) {
df <- as.data.frame(fit2[[ind]])
df$name <- gnames[ind]
df
})
time <- est <- event <- group <- NULL
df <- do.call(rbind, fit2_list)
df$event <- sapply(strsplit(df$name, split = gsep), `[`,
2)
df$group <- sapply(strsplit(df$name, split = gsep), `[`,
1)
df$std <- std <- sqrt(df$var)
pl <- ggplot(df, aes(time, est, color = event))
if (multiple_panels) {
pl <- ggplot(df, aes(time, est, color = event)) facet_wrap(~group)
}
else {
pl <- ggplot(df, aes(time, est, color = event, linetype = group))
}
if (conf.int) {
pl <- pl geom_errorbar(aes(ymin = est - coef * std,
ymax = est coef * std), alpha = 0.2)
}
pl geom_line()
}
ggcompetingrisks_altered <- function (fit, gnames = NULL, gsep = " ", multiple_panels = TRUE,
ggtheme = theme_survminer(), coef = 1.96, conf.int = FALSE,
...)
{
stopifnot(any(class(fit) %in% c("cuminc", "survfitms")))
if (any(class(fit) == "cuminc")) {
pl <- ggcompetingrisks_cuminc_altered(fit = fit, gnames = gnames,
gsep = gsep, multiple_panels = multiple_panels,
coef = coef, conf.int = conf.int)
}
if (any(class(fit) == "survfitms")) {
pl <- ggcompetingrisks.survfitms(fit = fit)
}
pl <- pl ggtheme ylab("Probability of an event") xlab("Time")
ggtitle("Cumulative incidence functions")
ggpubr::ggpar(pl, ...)
}
ggcompetingrisks_altered(fit, multiple_panels = FALSE, conf.int = TRUE)
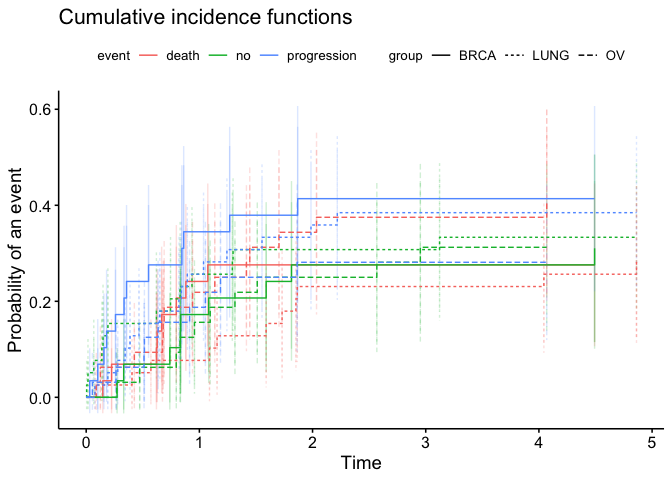
Created on 2021-11-29 by the reprex package (v2.0.1)
Is that the outcome you're after?
