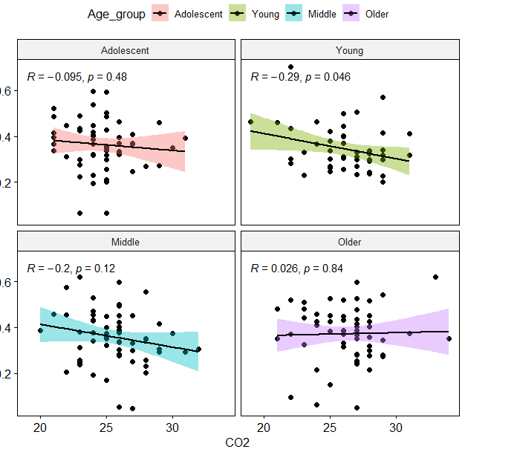
I have the following dataset:
structure(list(Age_group = structure(c(4L, 2L, 2L, 2L, 4L, 2L,
2L, 4L, 3L, 1L, 2L, 1L, 1L, 4L, 1L, 2L, 1L, 4L, 3L, 4L, 4L, 1L,
2L, 2L, 1L, 2L, 1L, 3L, 3L, 2L, 2L, 3L, 4L, 3L, 2L, 4L, 2L, 2L,
3L, 4L, 4L, 4L, 1L, 2L, 4L, 2L, 4L, 2L, 4L, 4L, 2L, 3L, 3L, 3L,
4L, 4L, 2L, 4L, 4L, 4L, 1L, 3L, 1L, 1L, 1L, 2L, 1L, 3L, 2L, 2L,
2L, 2L, 1L, 3L, 2L, 4L, 2L, 2L, 2L, 4L, 4L, 1L, 2L, 4L, 1L, 1L,
1L, 4L, 2L, 1L, 1L, 2L, 1L, 3L, 1L, 3L, 1L, 1L, 4L, 3L, 2L, 3L,
2L, 4L, 2L, 1L, 4L, 1L, 2L, 1L, 4L, 1L, 1L, 1L, 1L, 2L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 3L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 3L,
1L, 1L, 2L, 1L, 1L, 1L, 1L, 4L, 4L, 1L, 1L, 3L, 2L, 2L, 2L, 1L,
1L, 3L, 4L, 3L, 3L, 1L, 1L, 1L, 2L, 4L, 1L, 4L, 1L, 1L, 1L, 1L,
1L, 4L, 1L, 2L, 1L, 1L, 1L, 2L, 4L, 1L, 4L, 2L, 1L, 2L, 1L, 1L,
1L, 3L, 3L, 2L, 1L, 2L, 3L, 4L, 1L, 1L, 1L, 1L, 1L, 4L, 4L, 2L,
3L, 4L, 1L, 1L, 1L, 1L, 1L, 4L, 3L, 3L, 3L, 4L, 3L, 2L, 2L, 1L,
4L, 1L, 3L, 3L, 1L, 1L, 3L, 3L, 3L, 3L, 2L, 3L, 3L, 1L, 4L, 1L,
4L, 1L, 1L, 2L, 4L, 3L, 3L, 3L, 3L, 4L, 4L, 4L, 1L, 4L, 3L, 2L,
1L, 4L, 1L, 4L, 3L, 3L, 4L, 3L, 4L, 3L, 1L, 3L, 3L, 4L, 3L, 4L,
4L, 3L, 4L, 1L, 4L, 4L, 3L, 1L, 3L, 1L, 4L, 4L, 3L, 3L, 4L, 1L,
4L, 1L, 4L, 4L, 1L, 1L, 3L, 3L, 3L, 4L, 2L, 4L, 3L, 3L, 1L, 3L,
4L, 3L, 3L, 1L, 3L, 4L, 4L, 1L, 2L, 3L, 3L, 4L, 4L, 3L, 3L, 3L,
3L, 3L, 4L, 3L, 1L, 1L, 3L, 4L, 3L, 3L, 3L, 1L, 2L, 2L, 1L), .Label = c("Adolescent",
"Young", "Middle", "Older"), class = "factor"), Value = c(0.344845,
0.290967, 0.246231, 0.262066, 0.214854, 0.369023, 0.244076, 0.280915,
0.30564, 0.296507, 0.323117, 0.276703, 0.225361, 0.415376, 0.26803,
0.297092, 0.39287, 0.373648, 0.231434, 0.215282, 0.402466, 0.324974,
0.234958, 0.255247, 0.247927, 0.200748, 0.194252, 0.171439, 0.276834,
0.201723, 0.309028, 0.203337, 0.433123, 0.242758, 0.30205, 0.370564,
0.267963, 0.283591, 0.336721, 0.596052, 0.244396, 0.387599, 0.347128,
0.227341, 0.432896, 0.282985, 0.284935, 0.231549, 0.524341, 0.33092,
0.236906, 0.54037, 0.378644, 0.206526, 0.0978536, 0.252193, 0.332135,
0.315254, 0.280426, 0.217306, 0.23161, 0.240526, 0.446109, 0.41504,
0.28913, 0.269704, 0.317209, 0.433796, 0.392358, 0.299284, 0.338003,
0.311145, 0.426087, 0.339064, 0.506221, 0.519461, 0.464624, 0.422413,
0.311408, 0.384049, 0.30677, 0.316512, 0.274162, 0.426846, 0.437163,
0.350454, 0.406764, 0.502995, 0.330374, 0.299821, 0.43784, 0.329894,
0.48, 0.351307, 0.355426, 0.377012, 0.349693, 0.302153, 0.152044,
0.423236, 0.569011, 0.451337, 0.402483, 0.54266, 0.368916, 0.300246,
0.328711, 0.44537, 0.338924, 0.378004, 0.484292, 0.373512, 0.655633,
0.320122, 0.376306, 0.701183, 0.42354, 0.354544, 0.366982, 0.485444,
0.2711, 0.39679, 0.499632, 0.380856, 0.364726, 0.460057, 0.254963,
0.368593, 0.210968, 0.338162, 0.338745, 0.498087, 0.366381, 0.452842,
0.225168, 0.456962, 0.414057, 0.313421, 0.434526, 0.217877, 0.338147,
0.300099, 0.516165, 0.375086, 0.460186, 0.373398, 0.309855, 0.296928,
0.301164, 0.334937, 0.320049, 0.389919, 0.282245, 0.241675, 0.332736,
0.593453, 0.201379, 0.416399, 0.371206, 0.4048, 0.414817, 0.4947,
0.593219, 0.376317, 0.318016, 0.395748, 0.352561, 0.350144, 0.543684,
0.444405, 0.336287, 0.0667227, 0.325322, 0.379068, 0.391071,
0.37585, 0.476663, 0.464114, 0.461864, 0.415283, 0.458221, 0.400008,
0.38393, 0.285078, 0.237714, 0.361987, 0.426509, 0.317339, 0.294408,
0.619243, 0.34253, 0.329934, 0.355375, 0.46283, 0.407967, 0.242693,
0.51851, 0.317998, 0.323249, 0.448899, 0.360369, 0.459298, 0.484034,
0.27694, 0.487715, 0.434585, 0.605315, 0.494404, 0.256854, 0.351891,
0.231474, 0.413763, 0.410932, 0.365665, 0.511102, 0.365337, 0.527372,
0.400869, 0.24765, 0.369774, 0.350247, 0.530748, 0.461709, 0.428728,
0.303493, 0.573203, 0.498893, 0.280537, 0.387132, 0.594904, 0.425032,
0.370547, 0.535847, 0.397682, 0.372345, 0.305478, 0.193977, 0.362042,
0.453853, 0.383845, 0.359185, 0.349271, 0.248476, 0.404103, 0.333776,
0.433578, 0.317914, 0.36847, 0.394821, 0.254976, 0.436492, 0.596257,
0.331286, 0.299685, 0.063502, 0.469766, 0.403892, 0.447094, 0.471031,
0.458835, 0.248689, 0.479741, 0.277219, 0.294354, 0.450719, 0.32319,
0.481539, 0.489301, 0.301525, 0.310258, 0.415681, 0.42438, 0.320633,
0.441025, 0.0533728, 0.252189, 0.317907, 0.401426, 0.282361,
0.501992, 0.417136, 0.273503, 0.448618, 0.459488, 0.286582, 0.336108,
0.289597, 0.42585, 0.367346, 0.525273, 0.456723, 0.411294, 0.299206,
0.31401, 0.350646, 0.389548, 0.34972, 0.357895, 0.45329, 0.452023,
0.408471, 0.428022, 0.572826, 0.340292, 0.0470799, 0.326013,
0.38702, 0.375492, 0.555507, 0.403654, 0.620388, 0.259259, 0.386142,
0.389715, 0.305789, 0.39022, 0.385585, 0.0526119, 0.379378, 0.411465,
0.376643, 0.0645194, 0.519351, 0.459602, 0.520458), CO2 = c(29L,
28L, 25L, 25L, 28L, NA, 28L, 29L, 32L, NA, 28L, NA, NA, 27L,
28L, 29L, 31L, 31L, NA, 24L, 27L, NA, 27L, 26L, NA, 29L, 24L,
25L, 26L, 29L, NA, 28L, 26L, NA, 22L, 26L, 25L, 22L, NA, 27L,
NA, 26L, 25L, 29L, 26L, NA, NA, 23L, 27L, 26L, 28L, NA, 24L,
22L, 22L, 27L, 23L, 26L, 27L, 28L, 24L, NA, 22L, 21L, NA, 27L,
24L, 24L, NA, 28L, 25L, 26L, 25L, 26L, 27L, 26L, 24L, 25L, 27L,
25L, NA, 25L, NA, 28L, NA, 30L, 27L, NA, 23L, 25L, NA, 27L, NA,
28L, 25L, 26L, NA, NA, 25L, 26L, 29L, 26L, 26L, 29L, 26L, NA,
NA, 24L, NA, NA, NA, NA, NA, 26L, NA, 22L, NA, 25L, 27L, 25L,
29L, 26L, NA, 26L, 21L, NA, 25L, 26L, 25L, 28L, 29L, 26L, NA,
27L, 23L, NA, NA, NA, 23L, NA, 25L, 28L, 28L, NA, 29L, 30L, NA,
27L, 25L, 26L, 24L, NA, NA, NA, 27L, 25L, 25L, 24L, NA, NA, NA,
NA, 25L, NA, 25L, NA, NA, 27L, NA, 26L, 21L, 25L, 26L, 25L, NA,
NA, 27L, 19L, 26L, NA, NA, NA, 24L, 26L, 23L, NA, NA, 29L, 31L,
33L, NA, NA, NA, NA, NA, 27L, 22L, 31L, 25L, 26L, NA, NA, 21L,
23L, 23L, 27L, NA, 26L, 23L, 34L, 28L, 29L, 31L, 24L, 23L, NA,
NA, 25L, 27L, 27L, 25L, 24L, NA, 24L, 26L, 22L, 26L, NA, 26L,
24L, 24L, NA, 24L, 26L, 22L, 29L, 24L, 25L, 24L, 26L, 28L, NA,
NA, 28L, 26L, 22L, NA, 27L, 21L, 27L, NA, 26L, NA, 27L, 24L,
24L, 24L, 25L, NA, 24L, 23L, 21L, 28L, 29L, 25L, 26L, 23L, NA,
26L, 22L, 29L, 23L, 28L, 23L, 26L, 27L, NA, 24L, 27L, 25L, NA,
29L, NA, NA, NA, NA, NA, 25L, 24L, 25L, 21L, NA, 23L, 23L, 21L,
26L, 28L, NA, 22L, 28L, 24L, NA, NA, 24L, 27L, 23L, 27L, 25L,
28L, 26L, 23L, 28L, NA, 26L, NA, NA, 20L, 27L, 23L, NA, NA, 23L,
NA, 21L, 21L)), row.names = c(NA, -325L), class = c("tbl_df",
"tbl", "data.frame"))
I used the following code to calculate correlations between X and Y and facet by age groups:
library(tidyverse)
library(ggpubr)
p <- ggscatter(DF, x = "CO2", y = "Value",
fill = "Age_group",
add = "reg.line", conf.int = TRUE,
cor.coef = TRUE, cor.method = "pearson")
facet(p, facet.by = "Age_group", scales = "free")
Which gives me the partly-desired output:
However, I would like to correct for having 4 separate correlations calculated. Is this possible as a quick option within my code? Like stat_compare_means has p.adjust.method options that I can implement?
Or do I need to calculate this separately and paste into the charts?
CodePudding user response:
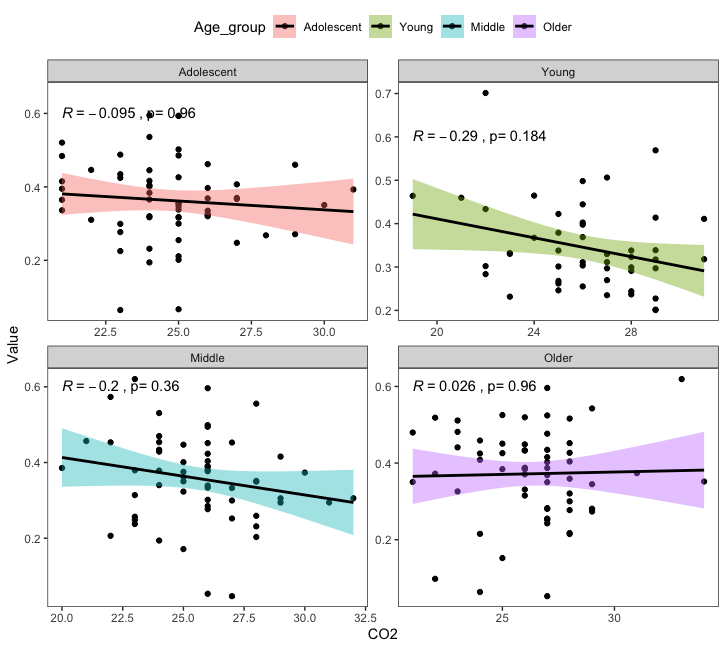
This is a bit hacky, but you you can manually adjust the p.value label with stats::p.adjust:
library(readr)
ggplot(DF, aes(x = CO2, y = Value, fill = Age_group, group = Age_group))
geom_point()
stat_smooth(method = "lm",
color = "black")
stat_cor(aes(label = paste0(..r.label..,
"~`,`~`p=`~",
p.adjust(readr::parse_number(..p.label..), n = 4))),
method = "pearson", label.y = 0.6)
facet_wrap(~Age_group, scales = "free")
theme_bw()
theme(panel.grid = element_blank(),
legend.position = "top")
Note that you will have to manually change the n= argument.