I have disease scores for a lot of trees with % dieback/ infection readings taken in two consecutive years. I can create a histogram in ggplot2 in r for each year, but how do I create a side-by-side bar plot showing the readings for each year side by side with ranges e.g. 0-10, 10-20, 20-30% up to 100% infection on the x axis?
I create a simple dataframe with two readings for each year:
diseaseyear1 <- c(20, 30, 40, 50, 30, 20, 60, 85, 45, 32, 20, 40)
diseaseyear2 <- c(30, 35, 42, 45, 25, 70, 65, 90, 40, 25, 35, 50)
totaldisease <- tibble(diseaseyear1, diseaseyear2)
totaldisease
#I can plot year 1
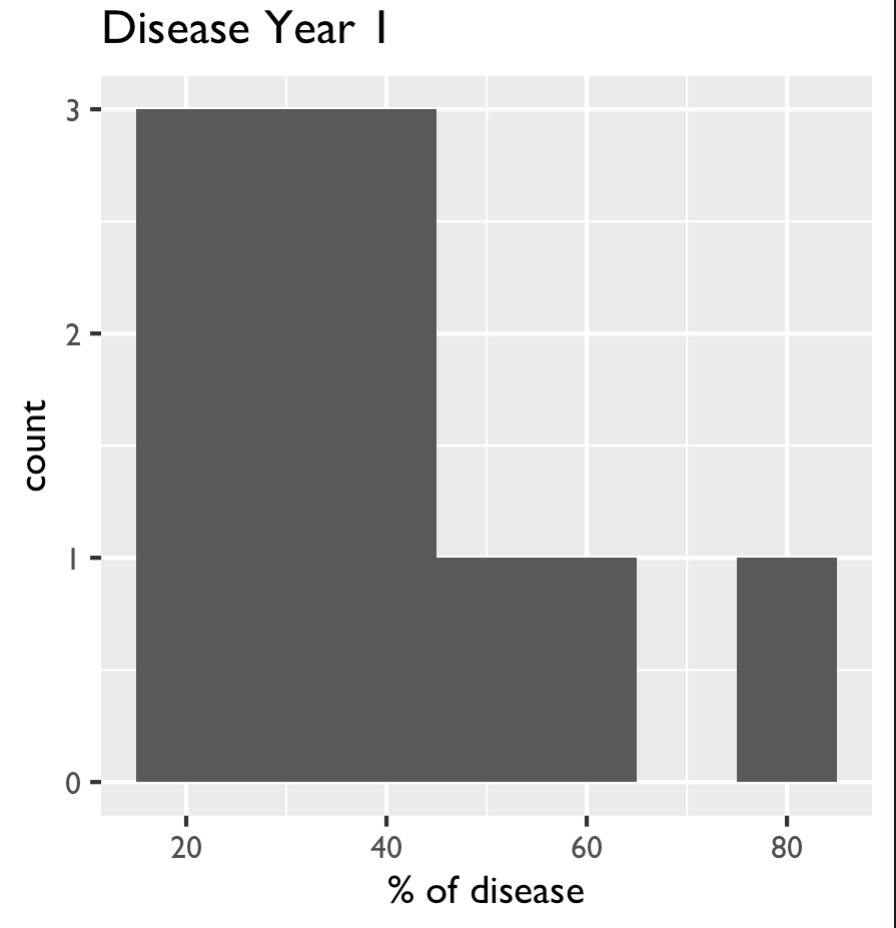
quartz(10, 5)
year1 <- ggplot(totaldisease)
aes(x=diseaseyear1)
geom_histogram(binwidth = 10)
labs(title = "Disease Year 1",
y = "count", x = "% of disease")
theme(text = element_text(size = 10, family = "GillSans"))
#I can also plot year 2
year1 <- ggplot(totaldisease)
aes(x=diseaseyear1)
geom_histogram(binwidth = 10)
labs(title = "Disease Year 1 & 2",
y = "count", x = "% of disease")
theme(text = element_text(size = 10, family = "GillSans"))
#but how can I combine these two histograms (or bar plots) as side by side bars in one chart?
CodePudding user response:
We need to first convert our data to a long format. This way, we can better plot the data.
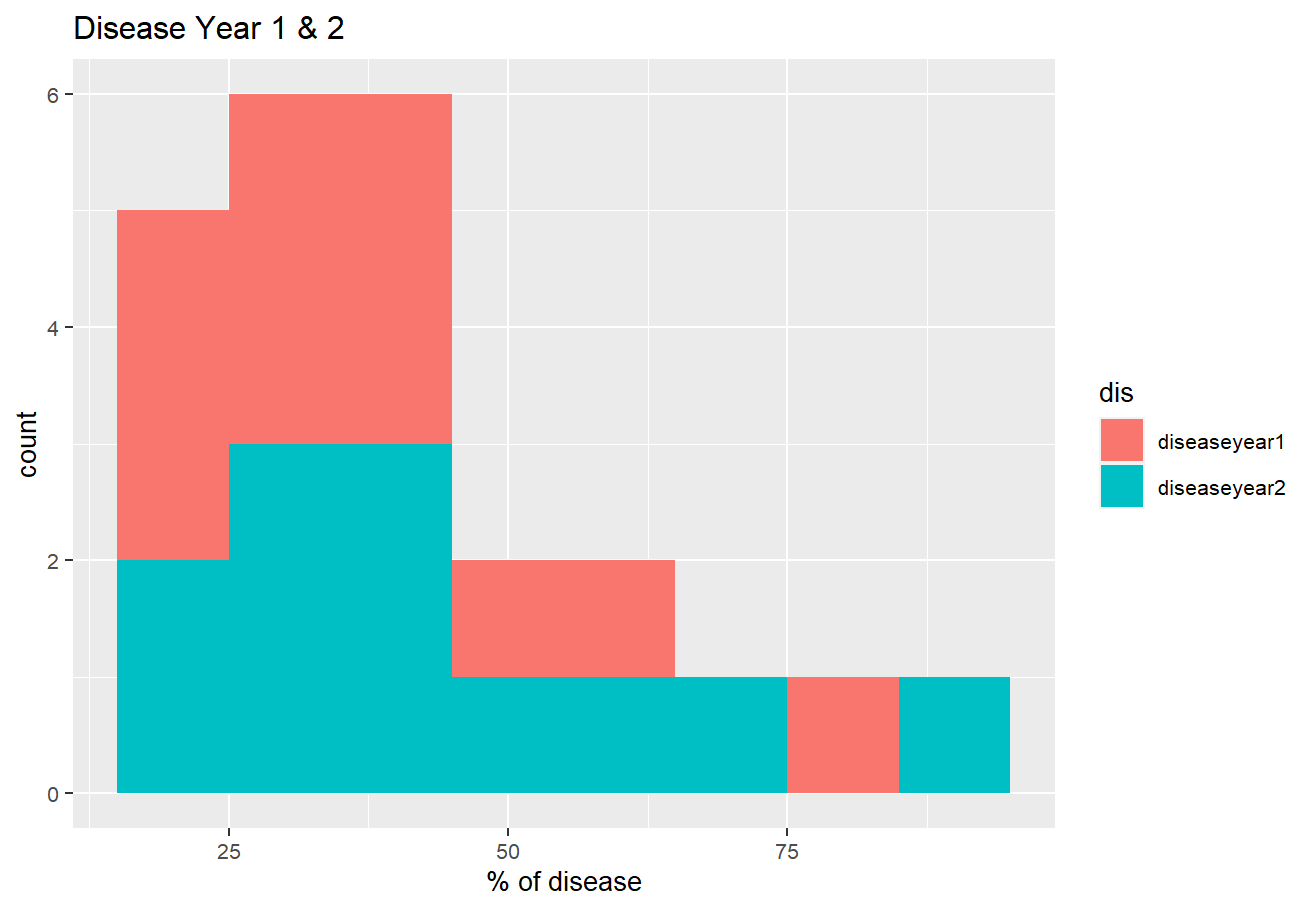
totaldisease %>%
tidyr::pivot_longer(names_to = "dis", values_to = "val",
cols = 1:ncol(.)) %>%
ggplot(aes(val, fill=dis))
geom_histogram(binwidth = 10)
labs(title = "Disease Year 1 & 2",
y = "count", x = "% of disease")
theme(text = element_text(size = 10, family = "GillSans"))
Result
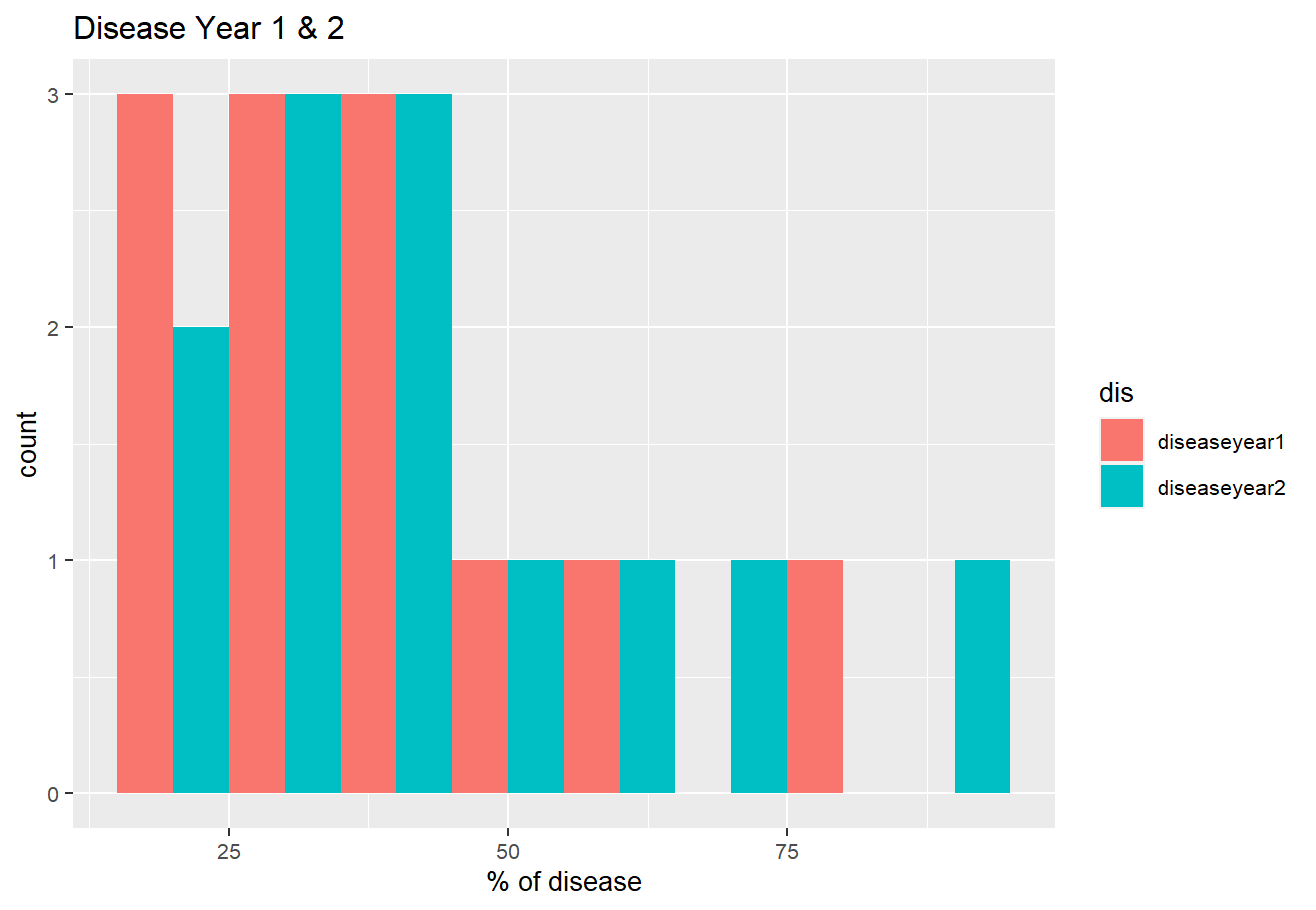
If you need it dodged
totaldisease %>%
tidyr::pivot_longer(names_to = "dis", values_to = "val",
cols = 1:ncol(.)) %>%
ggplot(aes(val, fill=dis))
geom_histogram(binwidth = 10, position = "dodge")
labs(title = "Disease Year 1 & 2",
y = "count", x = "% of disease")
theme(text = element_text(size = 10, family = "GillSans"))