I need to simulate a dataframe with 3 columns in R.
The first two columns must be two categorical variables, while the last one a continuous one.
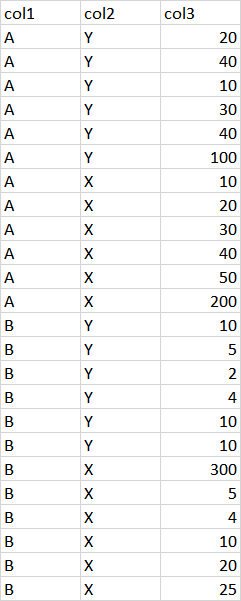
The dataframe should look like the following:
Numbers in col3 are just illustrative. What is important is that col1 (i.e., the first categorical variable) should present two possible values (i.e., "A" and "B" in my example), while col2 (i.e., the second categorical variable) should present other two possible values (i.e., "Y" and "X" in my example). As you can see, in this way we can identify four groups of rows:
- rows with "A" in col1 and "Y" in col2;
- rows with "A" in col1 and "X" in col2;
- rows with "B" in col1 and "Y" in col2;
- rows with "B" in col1" and "X" in col2.
What I want is that col3 is compiled in order to have outliers in each of the above four groups (in the sense that, for example, an outlier in the first group is not necessarily an outlier among the data included in the second).
CodePudding user response:
Not really sure about the distribution that you want for col3, but by using the normal distribution with the inputs defined by col1 and col2 and then codify them as a factor, you can do something like this:
nsim <- 1000
col1 <- sample(1:2, nsim, replace = T)
col2 <- sample(26:25, nsim, replace = T)
col3 <- rnorm(nsim, col1*col2, col2/col1)
col1 <- factor(col1, labels = LETTERS[1:2])
col2 <- factor(col2, labels = LETTERS[26:25])
df <- data.frame(col1, col2, col3)
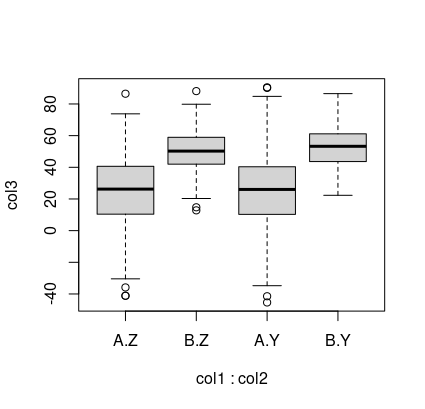
with(df, boxplot(col3~col1*col2))
If you want to specify each distribution (if there are not a large ammount), you can also use the ifelse function:
nsim <- 1000
col1 <- sample(LETTERS[1:2], nsim, replace = T)
col2 <- sample(LETTERS[26:25], nsim, replace = T)
df <- data.frame(col1, col2)
df$col3 <- apply(df, 1, function(y) ifelse(y[1] == "A", ifelse(y[2] == "Z", rnorm(1, 10, 2), rnorm(1, 11, 2)),
ifelse(y[2] == "Z", rnorm(1, 15, 2), rnorm(1, 19, 2))))
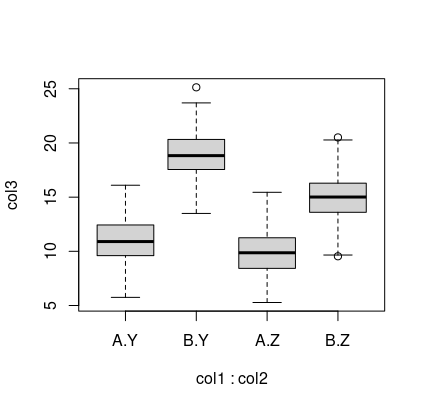
with(df, boxplot(col3~col1*col2))
CodePudding user response:
I would set up a function (generateDataset below) to generate the 4 datasets that you need individually. You could then join them together to get your full table.
I included nOutlier and scaleValue parameters in the function parameters. These allow you to insert outliers in your otherwise normal data.
library(tidyverse)
generateDataset <- function(col1Val, col2Val, meanVal, sdVal, nVal, nOutliers, scaleValue){
x <- data.frame(
col1 = rep(col1Val, 10),
col2 = rep(col2Val, 10),
col3 = rnorm(mean = meanVal, sd = sdVal, n=nVal)
)
y <- x %>%
sample_n(size = nOutliers) %>%
mutate(col3New = col3 * scaleValue)
z <- x %>%
anti_join(y, by=c("col1","col2","col3")) %>%
bind_rows(y %>% mutate(col3 = col3New) %>% select(-col3New))
return(z)
}
df <- generateDataset("A", "X", 10, 1, 200, 5, 2) %>%
bind_rows(generateDataset("A", "Y", 10, 4, 200, 1, 10)) %>%
bind_rows(generateDataset("B", "X", 20, 2, 200, 10, 15))
ggplot(df)
geom_boxplot(aes(x=col1, y=col3, colour=col2))