I found that it may be a problem with PyCharm's cache.
After I learned the sin function, I changed sin directly to cos and ran it without saving. The 2000th time was still the wrong result.
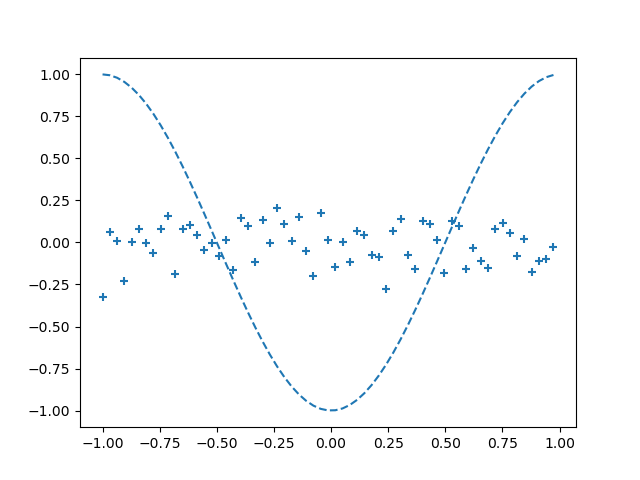
Epoch:0/2001 Error:0.2798077795267396
Epoch: 200/2001 Error: 0.27165245260858123
Epoch: 400/2001 Error: 0.2778566883056528
Epoch: 600/2001 Error: 0.26485675644837514
Epoch: 800/2001 Error: 0.2752758904739536
Epoch: 1000/2001 Error: 0.2633888652172328
Epoch: 1200/2001 Error: 0.2627593240503436
Epoch: 1400/2001 Error: 0.27195552955032104
Epoch: 1600/2001 Error: 0.27268507931720914
Epoch: 1800/2001 Error: 0.2689462168186385
Epoch: 2000/2001 Error: 0.2737487268797401
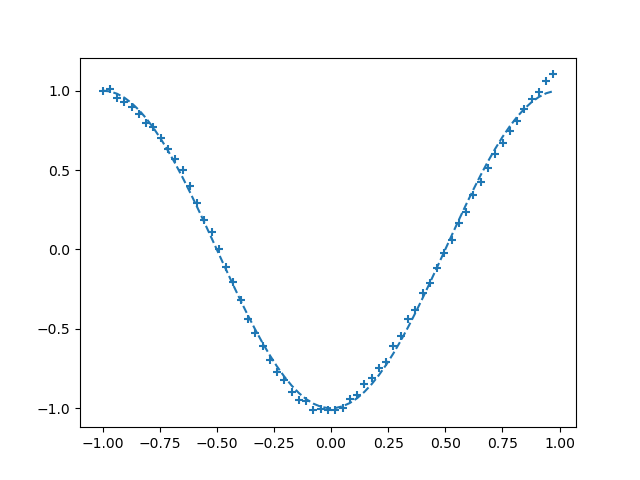
The 2000th results are as follows:
 But if I save and use "Reload all from disk", the error of the 400th time is already very small.
But if I save and use "Reload all from disk", the error of the 400th time is already very small.
Epoch:0/2001 Error:0.274032588963002
Epoch: 200/2001 Error: 0.2718715689675884
Epoch: 400/2001 Error: 0.0014035324029329518
Epoch:600/2001 Error:0.0004188502356206808
Epoch: 800/2001 Error: 0.000202233202030069
Epoch: 1000/2001 Error: 0.00014405423567078488
Epoch: 1200/2001 Error: 0.00011676179819916471
Epoch: 1400/2001 Error: 0.00011185491417278027
Epoch: 1600/2001 Error: 0.000105762467718704
Epoch:1800/2001 Error:8.768434766422346e-05
Epoch: 2000/2001 Error: 9.686019331806035e-05
The 400th results are as follows:
I use neural network back propagation regression to learn the cos function. When I learn the sin function, it is normal. If it is changed to cos, it is abnormal. What is the problem?
correct_data = np.cos(input_data)
Related settings:
1.The activation function of the middle layer: sigmoid function
2.Excitation function of the output layer: identity function
3.Loss function: sum of squares error
4.Optimization algorithm: stochastic gradient descent method
5.Batch size: 1
My code is as follows:
import numpy as np
import matplotlib.pyplot as plt
# - Prepare to input and correct answer data -
input_data = np.arange(0, np.pi * 2, 0.1) # input
correct_data = np.cos(input_data) # correct answer
input_data = (input_data - np.pi) / np.pi # Converge the input to the range of -1.0-1.0
n_data = len(correct_data) # number of data
# - Each setting value -
n_in = 1 # The number of neurons in the input layer
n_mid = 3 # The number of neurons in the middle layer
n_out = 1 # The number of neurons in the output layer
wb_width = 0.01 # The spread of weights and biases
eta = 0.1 # learning coefficient
epoch = 2001
interval = 200 # Display progress interval practice
# -- middle layer --
class MiddleLayer:
def __init__(self, n_upper, n): # Initialize settings
self.w = wb_width * np.random.randn(n_upper, n) # weight (matrix)
self.b = wb_width * np.random.randn(n) # offset (vector)
def forward(self, x): # forward propagation
self.x = x
u = np.dot(x, self.w) self.b
self.y = 1 / (1 np.exp(-u)) # Sigmoid function
def backward(self, grad_y): # Backpropagation
delta = grad_y * (1 - self.y) * self.y # Differentiation of Sigmoid function
self.grad_w = np.dot(self.x.T, delta)
self.grad_b = np.sum(delta, axis=0)
self.grad_x = np.dot(delta, self.w.T)
def update(self, eta): # update of weight and bias
self.w -= eta * self.grad_w
self.b -= eta * self.grad_b
# - Output layer -
class OutputLayer:
def __init__(self, n_upper, n): # Initialize settings
self.w = wb_width * np.random.randn(n_upper, n) # weight (matrix)
self.b = wb_width * np.random.randn(n) # offset (vector)
def forward(self, x): # forward propagation
self.x = x
u = np.dot(x, self.w) self.b
self.y = u # Identity function
def backward(self, t): # Backpropagation
delta = self.y - t
self.grad_w = np.dot(self.x.T, delta)
self.grad_b = np.sum(delta, axis=0)
self.grad_x = np.dot(delta, self.w.T)
def update(self, eta): # update of weight and bias
self.w -= eta * self.grad_w
self.b -= eta * self.grad_b
# - Initialization of each network layer -
middle_layer = MiddleLayer(n_in, n_mid)
output_layer = OutputLayer(n_mid, n_out)
# -- learn --
for i in range(epoch):
# Randomly scramble the index value
index_random = np.arange(n_data)
np.random.shuffle(index_random)
# Used for the display of results
total_error = 0
plot_x = []
plot_y = []
for idx in index_random:
x = input_data[idx:idx 1] # input
t = correct_data[idx:idx 1] # correct answer
# Forward spread
middle_layer.forward(x.reshape(1, 1)) # Convert the input to a matrix
output_layer.forward(middle_layer.y)
# Backpropagation
output_layer.backward(t.reshape(1, 1)) # Convert the correct answer to a matrix
middle_layer.backward(output_layer.grad_x)
# Update of weights and biases
middle_layer.update(eta)
output_layer.update(eta)
if i % interval == 0:
y = output_layer.y.reshape(-1) # Restore the matrix to a vector
# Error calculation
total_error = 1.0 / 2.0 * np.sum(np.square(y - t)) # Square sum error
# Output record
plot_x.append(x)
plot_y.append(y)
if i % interval == 0:
# Display the output with a graph
plt.plot(input_data, correct_data, linestyle="dashed")
plt.scatter(plot_x, plot_y, marker=" ")
plt.show()
# Display the number of epochs and errors
print("Epoch:" str(i) "/" str(epoch), "Error:" str(total_error / n_data))
CodePudding user response:
If increasing the number of epochs worked, the model needed more training.
But you may be overfitting... Notice that the cosine function is a periodic function, yet you are using only monotonic functions (sigmoid, and identity) to approximate it.
So while on the bounded interval of your data it may work:
It does not generalize well: