I am generating histograms using the plotly package in R to interactively inspect the distribution. In some cases, I have to apply a logarithmic function to make the plot easier to interpret. However, and as you would expect, when using plotly, the log-scaled value is presented in the tooltip box. What I would like to achieve is for the original values to be placed in the tooltip box instead of the log scale values.
Here is some example data and code:-
library(tidyverse)
library(plotly)
df <- data.frame(
sex=factor(rep(c("F", "M"), each=200)),
weight=round(c(rnorm(200, mean=55, sd=5),
rnorm(200, mean=65, sd=5)))
)
p <- ggplot(df, aes(x=weight))
geom_histogram()
scale_x_log10()
ggplotly(p)
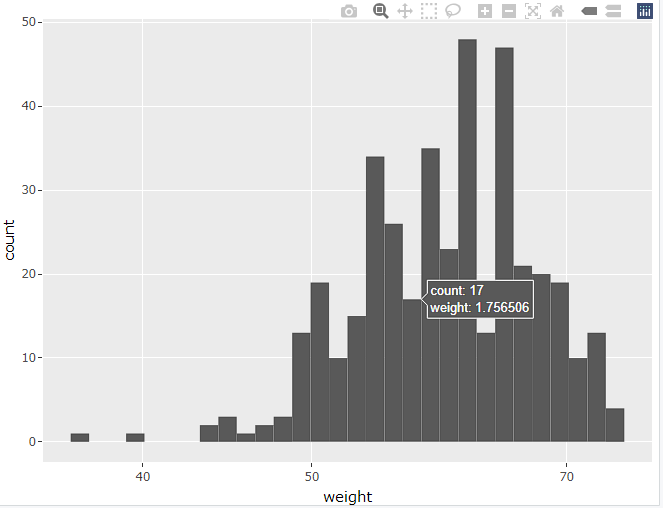
Which gives you this outcome:-
Can someone point me in the right direction? Thanks!
CodePudding user response:
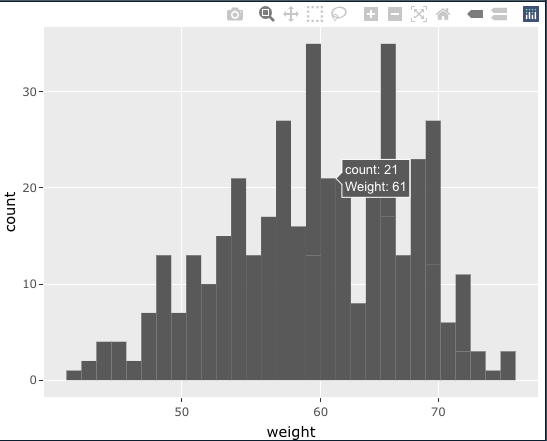
You can paste your weight in ggplot(aes(text = )), and only include text and y in ggplotly(tooltip = c("y", "text")).
p <- ggplot(df, aes(x = weight, text = paste("Weight:", weight)))
geom_histogram()
scale_x_log10()
ggplotly(p, tooltip = c("y", "text"))