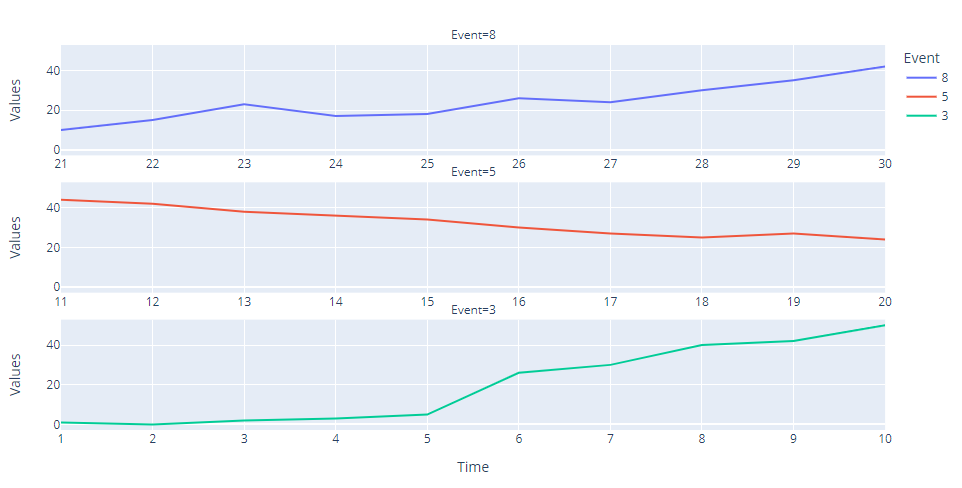
I have a dataframe which has some values against the time data. Here the time data is in discrete units as int datatype is given in the Time column. In this dataframe the time intervals are irregular, describing some event which is given in the Event column. And the values that must be plotted are in Values column.
data = {'Time':[21,22,23,24,25,26,27,28,29,30,11,12,13,14,15,16,17,18,19,20,1,2,3,4,5,6,7,8,9,10],
'Event':[8,8,8,8,8,8,8,8,8,8,5,5,5,5,5,5,5,5,5,5,3,3,3,3,3,3,3,3,3,3],
'Values':[10,15,23,17,18,26,24,30,35,42,44,42,38,36,34,30,27,25,27,24,1,0,2,3,5,26,30,40,42,50]}
data_df = pd.DataFrame(data)
data_df
With the following code, I was able to get the graphs seperated by events against their time values:
fig = px.line(data_df,
x='Time',
y='Values',
color='Event',
facet_col = 'Event',
facet_col_wrap=1
)
fig.update_xaxes(matches=None, showticklabels = True)
fig.show()
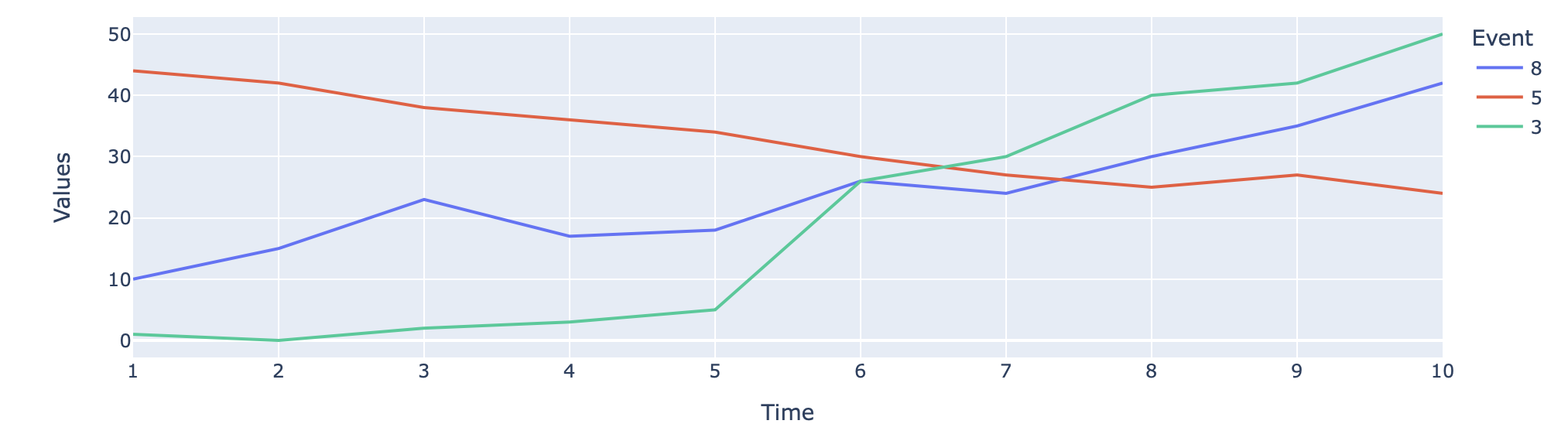
Now, I am curious if it is possible to somehow bring these graphs in a single graph with normalised x-axis. Lowerlimit of time value = 0. Upperlimit of timevalue = maximum of time duations of all the events. Basically x-axis should be large enough to accomodate all graphs.
Desired Output:
 The method I can think of is to create another column with normalised
The method I can think of is to create another column with normalised Time values and then plotting a single graph. Here I am curious to see if that can be avoided. Any information regarding this is appreciated.
CodePudding user response:
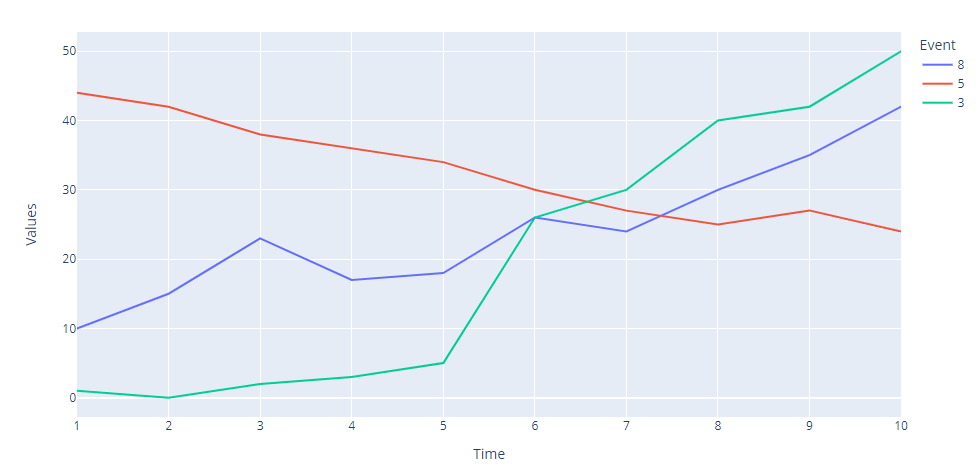
- have used pandas to rebase Time to be a sequence for each Event
- plotly is then straight forward
import pandas as pd
import plotly.express as px
# fmt: off
data = {'Time':[21,22,23,24,25,26,27,28,29,30,11,12,13,14,15,16,17,18,19,20,1,2,3,4,5,6,7,8,9,10],
'Event':[8,8,8,8,8,8,8,8,8,8,5,5,5,5,5,5,5,5,5,5,3,3,3,3,3,3,3,3,3,3],
'Values':[10,15,23,17,18,26,24,30,35,42,44,42,38,36,34,30,27,25,27,24,1,0,2,3,5,26,30,40,42,50]}
# fmt: on
data_df = pd.DataFrame(data)
px.line(
data_df.groupby("Event").apply(lambda d: d.assign(Time=range(1, len(d) 1, 1))),
x="Time",
y="Values",
color="Event",
)