I want to represent subclusters within clusters on variables using line graphs. I am using R.
I have two categorical variables (clusters; denoted below as a,b,c) which are nested such that each cluster has multiple subclusters (a1, a2, a3, b1, b2 and so on) within it.
I also have multiple numeric variables which I want to display by cluster and subcluster. I would like to use line graphs to display the means of the numeric variables. I have succeeded in displaying the means using summarize(),melt(), and ggplot with facet_wrap to separate the clusters. However, I don't know how I could display the subclusters.
I want to display the cluster means in with a thick black line, while displaying the subcluster means on the same graph, but "greyed out" and thinner to de-emphasize them. I have successfully used facet_wrap to separate the clusters, but I cannot think of how to get the subcluster means on the same graph.
I generated this dataset to illustrate the issue:
library(reshape)
library(tidyverse)
library(reshape)
library(tidyverse)
cases <- c(1:27)
cluster1 <- c(
"a","a","a","a","a","a","a","a","a",
"b","b","b","b","b","b","b","b","b",
"c","c","c","c","c","c","c","c","c")
cluster2 <- c(
"a1","a1","a1",
"a2","a2","a2",
"a3","a3","a3",
"b1","b1","b1",
"b2","b2","b2",
"b3","b3","b3",
"c1","c1","c1",
"c2","c2","c2",
"c3","c3","c3")
v1 <- runif(27,min = -2, max = 2)
v2 <- runif(27,min = -3, max = 1)
v3 <- runif(27,min = -4, max = 0)
df <- data.frame(cases,cluster1,cluster2,v1,v2,v3)
means.df <- subset(df) %>%
group_by(cluster1)%>%
summarise_at(vars(c(3:5)),mean)
means.df <- as.data.frame(means.df)
melt.df <- melt(means.df,id ="cluster1")
ggplot(data = melt.df,aes(x = variable, y = value, group = cluster1))
geom_line()
geom_point()
ylab("Mean")
theme(axis.text.x = element_text(angle = 90,hjust = 1,vjust=0.3))
facet_wrap(facets="cluster1")
Thank you in advance. Please let me know if I can provide more details.
CodePudding user response:
You could achieve your desired result by
- Creating a dataframe with the means by cluster1 and cluster2
- Passing this dataframe to the
dataargument of a secondgeom_line
library(reshape)
library(tidyverse)
set.seed(123)
means.df <- df %>%
group_by(cluster1) %>%
summarise(across(starts_with("v"), mean)) %>%
as.data.frame()
melt.df <- melt(means.df, "cluster1")
means.df2 <- df %>%
group_by(cluster1, cluster2) %>%
summarise(across(starts_with("v"), mean))%>%
as.data.frame()
melt.df2 <- melt(means.df2, c("cluster1", "cluster2"))
ggplot(data = melt.df, mapping = aes(x = variable, y = value, group = cluster1))
geom_line(data = melt.df2, aes(group = cluster2), color = "grey", alpha = .6)
geom_line(color = "black")
geom_point()
ylab("Mean")
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 0.3))
facet_wrap(facets = "cluster1")
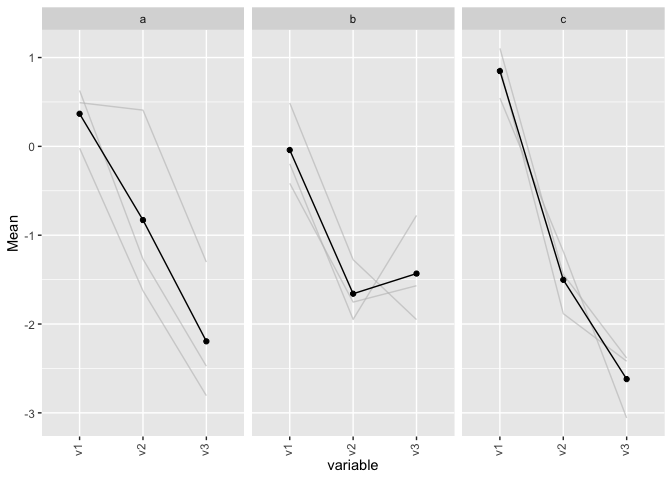
CodePudding user response:
You can use a different data origin to plot different layers of data as in geom_line(data = plotU, ...)
df <- data.frame(cases,cluster1,cluster2,v1,v2,v3) |>
pivot_longer(cols = starts_with("v"), names_to = "variable")
plotD <- df |>
group_by(variable, cluster1, cluster2) |>
summarize(value = mean(value), .groups = "drop")
plotU <- df |>
group_by(variable, cluster1) |>
summarize(value = mean(value), .groups= "drop")
ggplot(plotD, aes(x = variable, y = value))
geom_line(aes(group = cluster2), alpha = 0.5, lty = 2)
geom_line(data = plotU, color = "black", group = "cluster1", size = 1.4)
ylab("Mean")
theme(axis.text.x = element_text(angle = 90,hjust = 1,vjust=0.3))
facet_wrap(facets="cluster1")