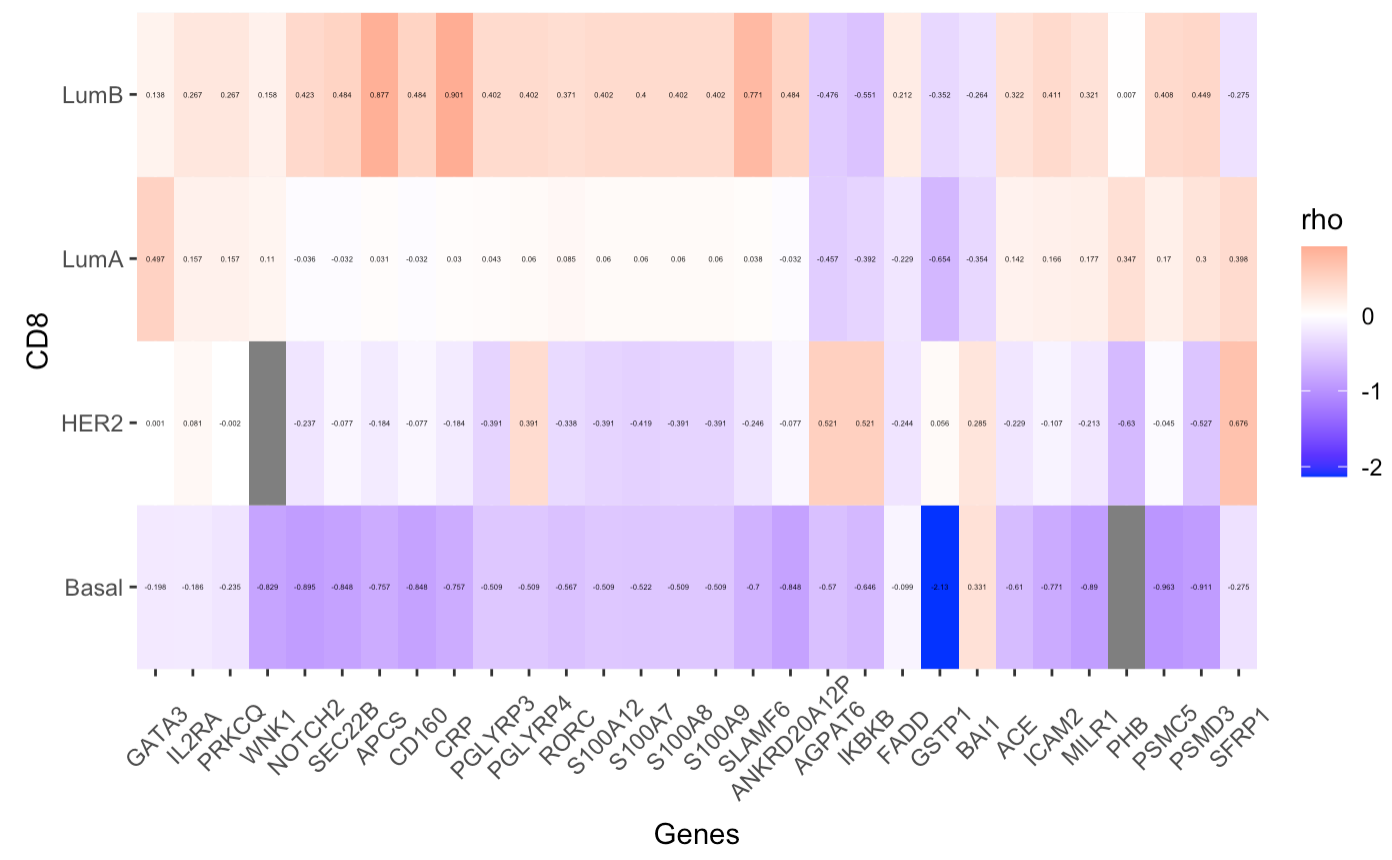
I need your help! I am plotting a heatmap and I do not have any error, but I do not know how to resolve it because I haven't seen any similar post. I have 2 dataframes:
DF1:
Basal HER2 LumA LumB
GATA3 -0.198 0.001 0.497 0.138
IL2RA -0.186 0.081 0.157 0.267
PRKCQ -0.235 -0.002 0.157 0.267
WNK1 -0.829 NA 0.110 0.158
NOTCH2 -0.895 -0.237 -0.036 0.423
SEC22B -0.848 -0.077 -0.032 0.484
APCS -0.757 -0.184 0.031 0.877
CD160 -0.848 -0.077 -0.032 0.484
CRP -0.757 -0.184 0.030 0.901
PGLYRP3 -0.509 -0.391 0.043 0.402
PGLYRP4 -0.509 0.391 0.060 0.402
RORC -0.567 -0.338 0.085 0.371
S100A12 -0.509 -0.391 0.060 0.402
S100A7 -0.522 -0.419 0.060 0.400
S100A8 -0.509 -0.391 0.060 0.402
S100A9 -0.509 -0.391 0.060 0.402
SLAMF6 -0.7 -0.246 0.038 0.771
ANKRD20A12P -0.848 -0.077 -0.032 0.484
AGPAT6 -0.570 0.521 -0.457 -0.476
IKBKB -0.646 0.521 -0.392 -0.551
FADD -0.099 -0.244 -0.229 0.212
GSTP1 -2.130 0.056 -0.654 -0.352
BAI1 0.331 0.285 -0.354 -0.264
ACE -0.610 -0.229 0.142 0.322
ICAM2 -0.771 -0.107 0.166 0.411
MILR1 -0.890 -0.213 0.177 0.321
PHB NA -0.630 0.347 0.007
PSMC5 -0.963 -0.045 0.170 0.408
PSMD3 -0.911 -0.527 0.300 0.449
SFRP1 -0.275 0.676 0.398 -0.275
DF2(p-values):
Basal HER2 LumA LumB
GATA3 0.544 0.974 0.514 0.629
IL2RA 0.640 0.758 0.985 0.521
PRKCQ 0.473 0.975 0.985 0.521
WNK1 0.055 NA 0.687 0.315
NOTCH2 0.023 0.584 0.707 0.927
SEC22B 0.031 0.948 0.703 0.851
APCS 0.127 0.900 0.369 0.201
CD160 0.031 0.948 0.703 0.851
CRP 0.127 0.900 0.385 0.126
PGLYRP3 0.245 0.379 0.350 0.206
PGLYRP4 0.245 0.379 0.281 0.206
RORC 0.166 0.502 0.203 0.431
S100A12 0.245 0.379 0.281 0.206
S100A7 0.225 0.379 0.281 0.262
S100A8 0.245 0.379 0.281 0.206
S100A9 0.245 0.379 0.281 0.206
SLAMF6 0.150 0.753 0.366 0.177
ANKRD20A12P 0.031 0.948 0.703 0.851
AGPAT6 0.233 0.060 0.006 0.050
IKBKB 0.156 0.060 0.024 0.018
FADD 0.448 0.393 0.037 0.489
GSTP1 0.200 1.000 0.002 0.438
BAI1 0.821 0.442 0.033 0.213
ACE 0.985 0.243 0.303 0.502
ICAM2 0.667 0.587 0.213 0.231
MILR1 0.503 0.330 0.276 0.492
PHB NA 0.025 0.053 0.847
PSMC5 0.424 0.776 0.212 0.278
PSMD3 0.436 0.035 0.215 0.131
SFRP1 0.510 0.013 0.021 0.510
and I've plotted a heatmap of one of them using this code:
require(reshape)
library(ggplot2)
CD8.m = melt(CD8)
colnames(CD8.m) <- c('Genes','CD8', 'rho')
ggplot(data = data.frame(CD8.m), aes(x = Genes, y = CD8, fill = rho))
geom_tile() scale_fill_gradient2(low = "blue", mid = "white", high = "red")
scale_x_discrete(expand = c(0, 0)) scale_y_discrete(expand = c(0, 0))
theme(axis.text.x = element_text(vjust=0.6, angle=45))
geom_text(aes(label = rho), color = "black", size = 1)
But now I would like to add the p-values below the label of geom_text in parenthesis, and if the p-value is <0.05, it should be bold.
Any help is more than wellcome! Thanks!!
CodePudding user response:
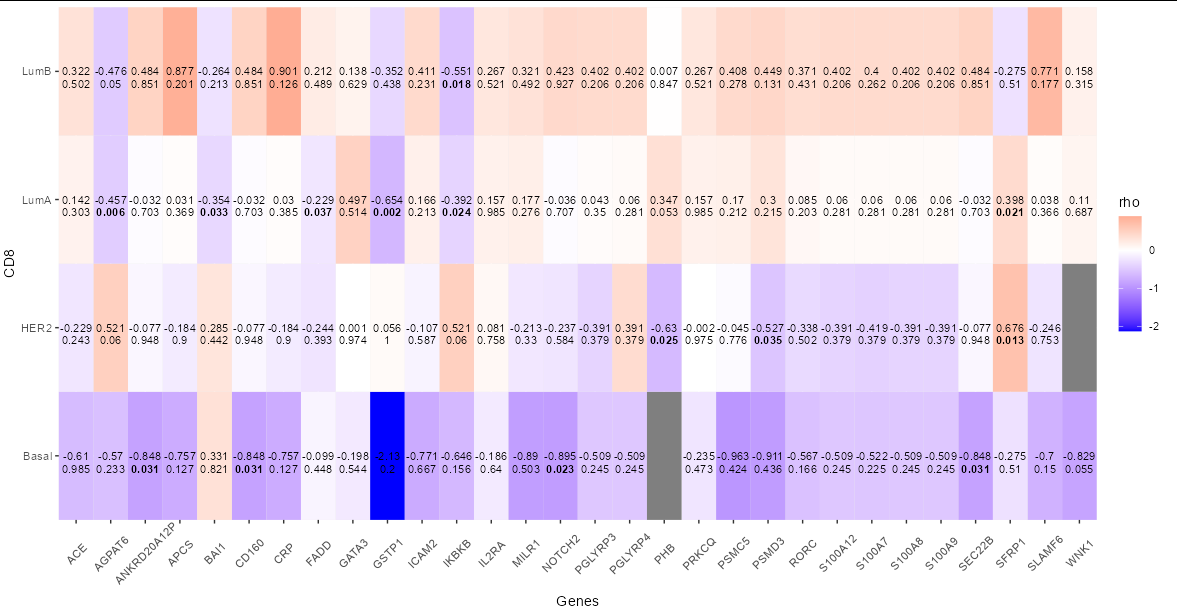
You need to convert DF2 into long format too. Then you can use the p values as the labels by passing DF2 as an argument to a different geom_text call. You can also map the fontface aesthetic to the the p values to get the bold font for "significant*" p values. Finally, you need to nudge the p value labels down a bit.
DF2.m <- melt(DF2)
colnames(DF2.m) <- c('Genes','CD8', 'pval')
ggplot(data = data.frame(CD8.m), aes(x = Genes, y = CD8))
geom_tile(aes(fill = rho))
scale_fill_gradient2(low = "blue", mid = "white", high = "red")
scale_x_discrete(expand = c(0, 0))
scale_y_discrete(expand = c(0, 0))
theme(axis.text.x = element_text(vjust = 0.6, angle = 45))
geom_text(aes(label = rho), color = "black", size = 3)
geom_text(data = DF2.m, aes(label = pval,
fontface = as.numeric(pval < 0.05) 1),
nudge_y = -0.1, size = 3)
A couple of points to note here are that firstly this plot is fairly illegible, and it may be best to find another way to present the data. Secondly, you are looking at 120 different p values, so we would expect about 6 of these p values to be less than 0.05 purely by chance. You might want to think about modifying your p value downwards using a Bonferroni correction or similar.