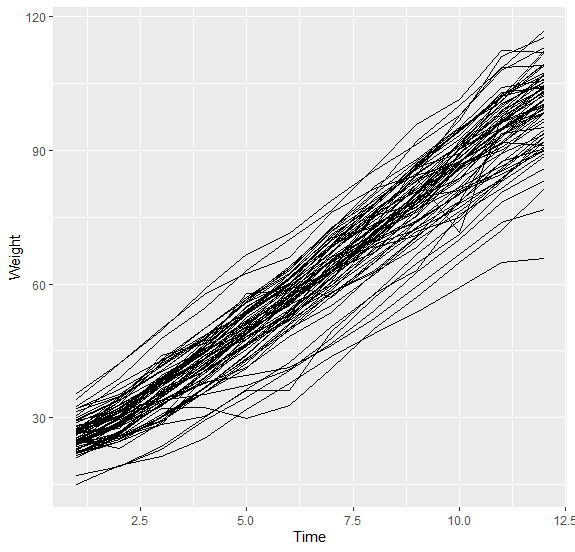
I am trying to create a custom spaghetti plot
Assuming this is my dataset
data(package = "geepack")
data("dietox")
head(dietox)
head(dietox, n =20)
Pig Evit Cu Litter Start Weight Feed Time
4601 Evit000 Cu000 1 26.50000 26.50000 NA 1
4601 Evit000 Cu000 1 26.50000 27.59999 5.200005 2
4601 Evit000 Cu000 1 26.50000 36.50000 17.600000 3
4601 Evit000 Cu000 1 26.50000 40.29999 28.500000 4
4601 Evit000 Cu000 1 26.50000 49.09998 45.200001 5
4601 Evit000 Cu000 1 26.50000 55.39999 56.900002 6
4601 Evit000 Cu000 1 26.50000 59.59998 71.700005 7
4601 Evit000 Cu000 1 26.50000 67.00000 86.800001 8
4601 Evit000 Cu000 1 26.50000 76.59998 104.900002 9
4601 Evit000 Cu000 1 26.50000 86.50000 123.000000 10
4601 Evit000 Cu000 1 26.50000 91.59998 140.900002 11
4601 Evit000 Cu000 1 26.50000 98.59998 160.000000 12
4602 Evit000 Cu035 1 28.29999 28.29999 NA 1
4602 Evit000 Cu035 1 28.29999 30.09999 3.300003 2
4602 Evit000 Cu035 1 28.29999 38.29999 13.200001 3
4602 Evit000 Cu035 1 28.29999 44.50000 26.100000 4
4602 Evit000 Cu035 1 28.29999 51.59998 43.600002 5
4602 Evit000 Cu035 1 28.29999 57.59998 55.200001 6
ggplot(data = dietox, aes(x = Time, y = Weight, group = Pig))
geom_line()
This creates a plot like this
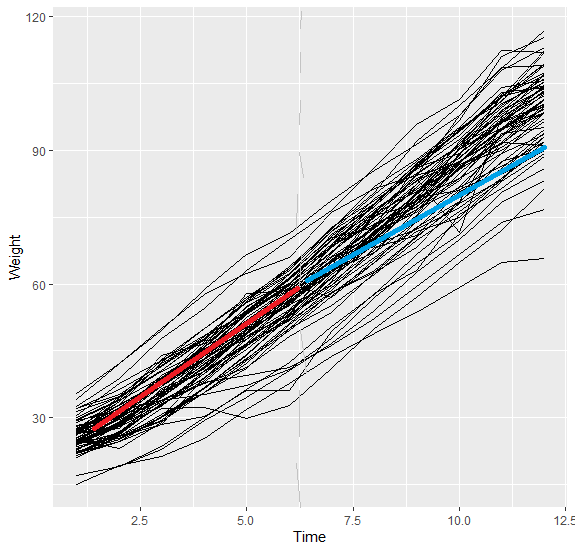
What I am interested in , is creating a gee predicted regression lines at two time points, 1)0- 6, 2) 6-12. The expected plot should look like this.
mf <- formula(Weight ~ Cu * (Time I(Time^2) I(Time^3)))
gee1 <- geeglm(mf, data=dietox, id=Pig, family="gaussian", corstr="ar1")
gee1
I would really appreciate your help.
CodePudding user response:
Your model takes the variable Cu into account, so each of your periods would have 3 different lines - one for each level of Cu. Since this is just sample data taken from the package, and you are looking for a single line in each period, I assume that your actual data is a function of a single variable.
The way to plot this is to manually predict lines for the two separate periods and plot them:
library(geepack)
library(ggplot2)
library(hrbrthemes)
data("dietox")
mf <- formula(Weight ~ Time I(Time^2) I(Time^3))
gee1 <- geeglm(mf, data = dietox[dietox$Time <= 6,],
id = Pig, family = "gaussian", corstr ="ar1")
gee2 <- geeglm(mf, data = dietox[dietox$Time >= 6,],
id = Pig, family = "gaussian", corstr ="ar1")
dietox$predictions <- numeric(1)
dietox$period <- "A"
dietox$predictions[dietox$Time <= 6] <- predict(gee1)
dietox$predictions[dietox$Time >= 6] <- predict(gee2)
dietox$period[dietox$Time >= 6] <- "B"
ggplot(data = dietox, aes(x = Time, y = Weight, group = Pig))
geom_line(aes(group = Pig), size = 2, alpha = 0.2)
geom_line(aes(y = predictions, group = interaction(Pig, period),
colour = period), size = 2)
theme_ipsum()
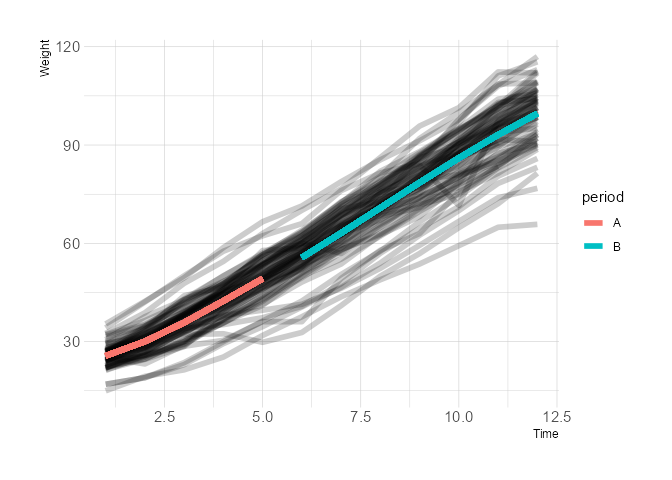
Created on 2022-03-09 by the reprex package (v2.0.1)