I am working with the R programming language.
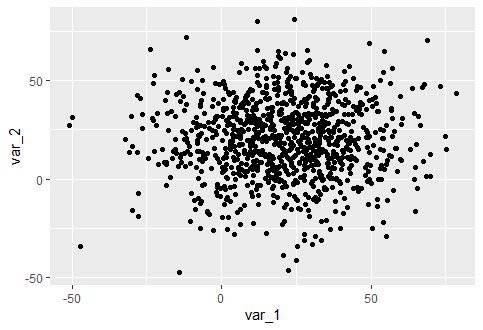
I generated some random data and plotted the following graph:
library(ggplot2)
var_1 = rnorm(1000,20,20)
var_2 = rnorm(1000,20,20)
my_data = data.frame(var_1, var_2)
ggplot(data=my_data, aes(x=var_1, y=var_2)) geom_point()
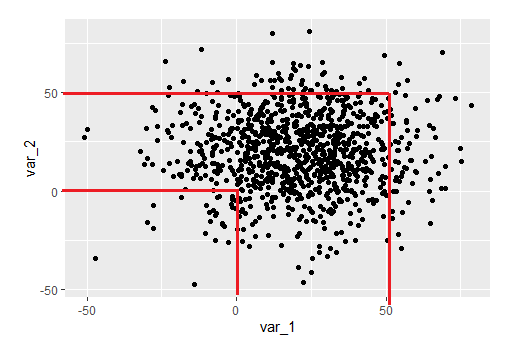
I now want to make the following pattern on this graph:
I tried to do this with the following code, but this is the closest I could get:
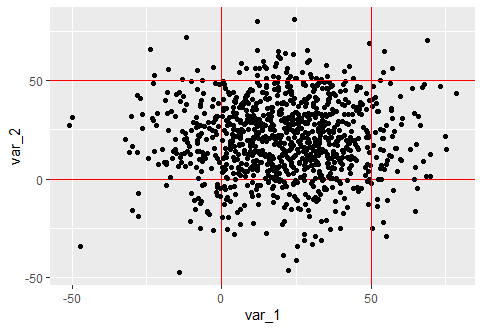
a1 = ggplot(data=my_data, aes(x=var_1, y=var_2))
geom_point()
geom_hline(yintercept=0, color = "red")
geom_vline(xintercept=0, color = "red")
a2 = a1 geom_point()
geom_hline(yintercept=50, color = "red")
geom_vline(xintercept=50, color = "red")
My Question: Could someone please show me a "direct" way to make this pattern on this graph? Is there some way to "erase" these extra lines?
Suppose I had a data frame ("my_points") that looked like this:
my_points = data.frame(var_1_point_1 = 0, var_1_point_2 = 50, var_2_point_1 = 0, var_2_point_2 = 50)
Could the graph be made using "my_data", and the lines be made by "referencing" the points in "my_points"? For example, something like this:
a1 = ggplot(data=my_data, aes(x=var_1, y=var_2))
geom_point()
geom_hline(yintercept my_points[1,2], color = "red")
geom_vline(xintercept=my_points[1,1], color = "red")
Thanks!
CodePudding user response:
You'll need to use geom_segment instead of geom_hline or geom_vline since you'll need to specify xend and yend for each segment.
library(tidyverse)
var_1 = rnorm(1000,20,20)
var_2 = rnorm(1000,20,20)
my_data = data.frame(var_1, var_2)
ggplot(data=my_data, aes(x=var_1, y=var_2))
geom_point()
geom_segment(aes(x = -50, xend = 0, y = 0, yend = 0), color = "red")
geom_segment(aes(x = 0, xend = 0, y = 0, yend = -50), color = "red")
geom_segment(aes(x = -50, xend = 50, y = 50, yend = 50), color = "red")
geom_segment(aes(x = 50, xend = 50, y = 50, yend = -50), color = "red")
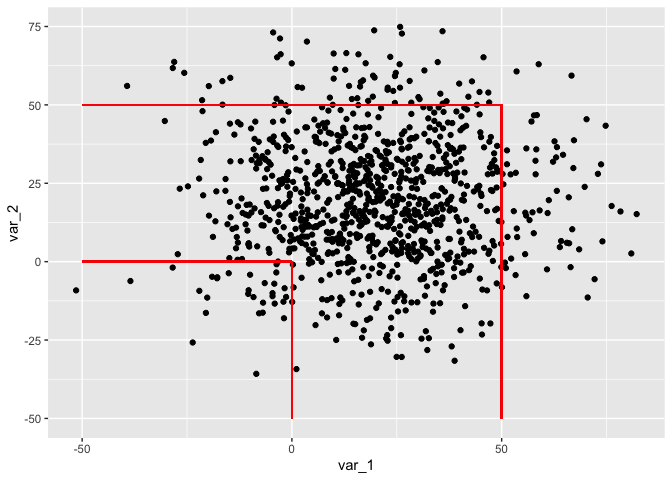
Created on 2022-03-21 by the reprex package (v2.0.1)
CodePudding user response:
People tend to forget about geom_linerange - in vanilla ggplot sadly only available for vertical lines. But there is also ggstance::geom_linerangeh. Those would allow you to pass a data frame with your x/y coordinates and you will "only" need two geom layers for vertical and for horizontal lines (if your lines always start at the edge of the plot). You could apply the same idea with geom_segment of course, then without need of another package.
library(ggplot2)
library(ggstance)
## changed data frame my points:
x <- y <- c(0, 50)
my_points <- data.frame(x, y)
ggplot()
geom_point(aes(x = var_1, y = var_2), data = my_data)
geom_linerange(aes(x = x, ymin = -Inf, ymax = y), data = my_points, color = "red", size = 2)
geom_linerangeh(aes(xmin = -Inf, xmax = x, y = y), data = my_points, color = "red", size = 2)
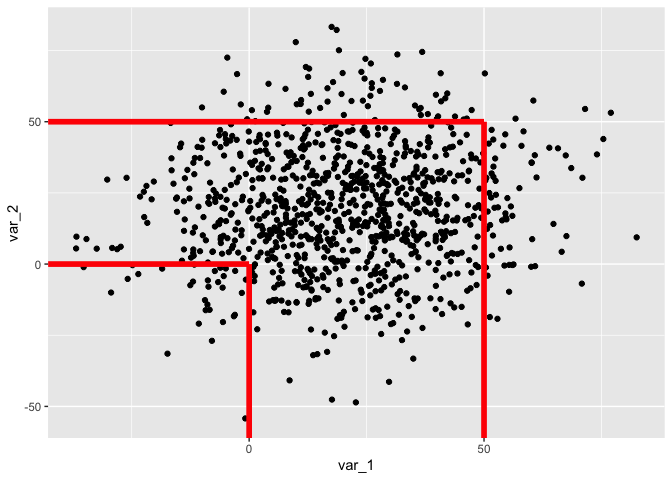
CodePudding user response:
Another option would be to make your own stat and use this with geom_segment:
library(ggplot2)
var_1 <- rnorm(1000, 20, 20)
var_2 <- rnorm(1000, 20, 20)
my_data <- data.frame(var_1, var_2)
## changed data frame my points:
x <- y <- c(0, 50)
my_points <- data.frame(x, y)
StatEdge <- ggproto("StatEdge", Stat, compute_group = function(data, scales) {
x <- c(rep(-Inf, nrow(data)), data$x)
y <- c(data$y, rep(-Inf, nrow(data)))
xend <- rep(data$x, nrow(data))
yend <- rep(data$y, nrow(data))
data.frame(x, xend, y, yend)
})
## now you can use the new stat for your geom
ggplot()
geom_point(aes(x = var_1, y = var_2), data = my_data)
geom_segment(data = my_points, aes(x, y), stat = "edge", size = 2, color = "red")
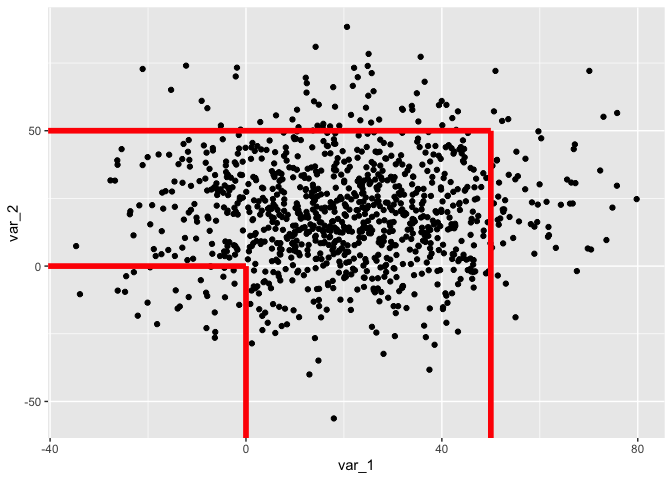
Created on 2022-03-21 by the reprex package (v2.0.1)