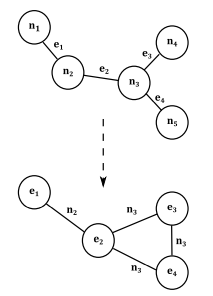
I am currently working with graph with labeled edges. The original adjacency matrix is a matrix with shape [n_nodes, n_nodes, n_edges] where each cell [i,j, k] is 1 if node i and j are connected via edge k.
I need to create a reverse of the original graph, where nodes become edges and edges become nodes, so i need a new matrix with shape [n_edges, n_edges, n_nodes], where each cell [i,j,k] is 1 if edges i and j have k as a common vertex.
The following code correctly completes the task, but the use of 5 nested for-loops is too slow, to process the amount of graphs with which I have to work seems to take about 700 hours.
Is there a better way to implement this?
n_nodes = extended_adj.shape[0]
n_edges = extended_adj.shape[2]
reversed_graph = torch.zeros(n_edges, n_edges, n_nodes, 1)
for i in range(n_nodes):
for j in range(n_nodes):
for k in range(n_edges):
#If adj_mat[i][j][k] == 1 nodes i and j are connected with edge k
#For this reason the edge k must be connected via node j to every outcoming edge of j
if extended_adj[i][j][k] == 1:
#Given node j, we need to loop through every other possible node (l)
for l in range(n_nodes):
#For every other node, we need to check if they are connected by an edge (m)
for m in range(n_edges):
if extended_adj[j][l][m] == 1:
reversed_graph[k][m][j] = 1
Thanks is advance.
CodePudding user response:
Compress the initial adjacency matrix from [Nn, Nn, Ne] to [Nn, Nn] by assigning the edge numbers directly to the matrix elements. This takes an exhaustive traversal of the initial matrix, taking O(Nn²Ne).
For every node n, you know the list of adjacent nodes with the corresponding edge index; so you generate all pairs of adjacent nodes ni, nj and write the node index n at the position (ei, ej). This takes O(Nn² Ne²) because you need to initialize the edges array.
Uncompress from [Ne, Ne] to [Ne, Ne, Nn] in time O(Ne²Nn).
Hence the total time is bounded by O(NnNe(Nn Ne)).
CodePudding user response:
Echoing the comments above, this graph representation is almost certainly cumbersome and inefficient. But that notwithstanding, let's define a vectorized solution without loops and that uses tensor views whenever possible, which should be fairly efficient to compute for larger graphs.
For clarity let's use [i,j,k] to index G (original graph) and [i',j',k'] to index G' (new graph). And let's shorten n_edges to e and n_nodes to n.
Consider the 2D matrix slice = torch.max(G,dim = 1). At each coordinate [a,b] of this slice, a 1 indicates that node a is connected by edge b to some other node (we don't care which).
slice = torch.max(G,dim = 1) # dimension [n,e]
We're well on our way to the solution, but we need an expression that tells us whether a is connected to edge b and another edge c, for all edges c. We can map all combinations b,c by expanding slice, copying it and transposing it, and looking for intersections between the two.
expanded_dim = [slice.shape[0],slice.shape[1],slice.shape[1]] # value [n,e,e]
# two copies of slice, expanded on different dimensions
expanded_slice = slice.unsqueeze(1).expand(expanded_dim) # dimension [n,e,e]
transpose_slice = slice.unsqueeze(2).expand(expanded_dim) # dimension [n,e,e]
G = torch.bitwise_and(expanded_slice,transpose_slice).int() # dimension [n,e,e]
G[i',j',k'] now equals 1 iff node i' is connected by edge j' to some other node, AND node i' is connected by edge k' to some other node. If j' = k' the value is 1 as long as one of the endpoints of that edge is i'.
Lastly, we reorder dimensions to get to your desired form.
G = torch.permute(G,(1,2,0)) # dimension [e,e,n]