I was wondering if anyone could shed some light into how I can average this data:
I have a .nc file with data (dimensions: 2029,64,32) which relates to time, latitude and longitude. Using these commands I can plot individual timesteps:
timestep = data.variables['precip'][0]
plt.imshow(timestep)
plt.colorbar()
plt.show()
Giving a graph in this format for the 0th timestep:
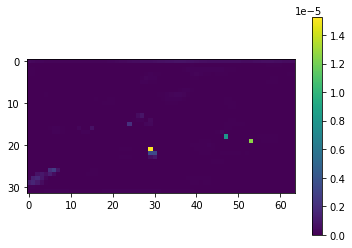
I was wondering if there was any way to average this first dimension (the snapshots in time).
CodePudding user response:
I think if you're using pandas and numpy this may help you.Look for more details
import pandas as pd
import numpy as np
data = np.array([10,5,8,9,15,22,26,11,15,16,18,7])
d = pd.Series(data)
print(d.rolling(4).mean())
CodePudding user response:
If you are looking to take a mean over all times, try using np.mean where you use the axis keyword to say which axis you want to average.
time_avaraged = np.mean(data.variables['precip'], axis = 0)
If you have NaN values then np.mean will give NaN for that lon/lat point. If you'd rather ignore them then use np.nanmean.
If you want to do specific times only, e.g. the first 1000 time steps, then you could do
time_avaraged = np.mean(data.variables['precip'][:1000,:,:], axis = 0)
