I'm currently in the process of creating a heatmap with plotly. Below is the sample dataset:
library(tidyverse)
library(plotly)
library(hrbrthemes)
set.seed(9999)
df <- data.frame(group.int = rep(c(rep("Prevention", 3), "Diagnosis", rep("Intervention", 2)), 6),
int = rep(c("Prevention 1", "Prevention 2", "Prevention 3", "Diagnosis 1", "Intervention 1", "Intervention 2"), 6),
group.outcome = c(rep("Efficacy", 12), rep("Safety", 18), rep("Cost-effectiveness", 6)),
outcome = c(rep("Efficacy 1", 6), rep("Efficacy 2", 6), rep("Safety 1", 6), rep("Safety 2", 6), rep("Safety 3", 6), rep("Cost-effectiveness 1", 6)),
n = sample(50:250, 36, rep = TRUE)
)
df$group.int <- factor(df$group.int, levels = c("Prevention", "Diagnosis", "Intervention"))
df$group.outcome <- factor(df$group.outcome, levels = c("Efficacy", "Safety", "Cost-effectiveness"))
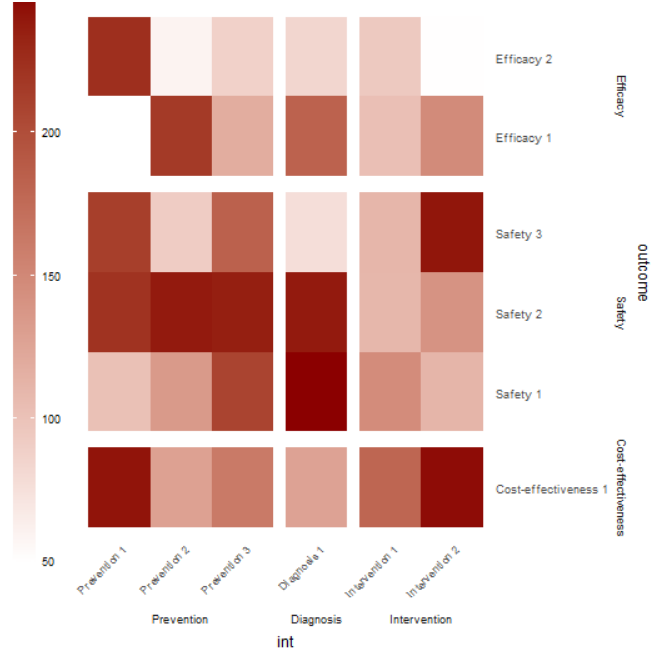
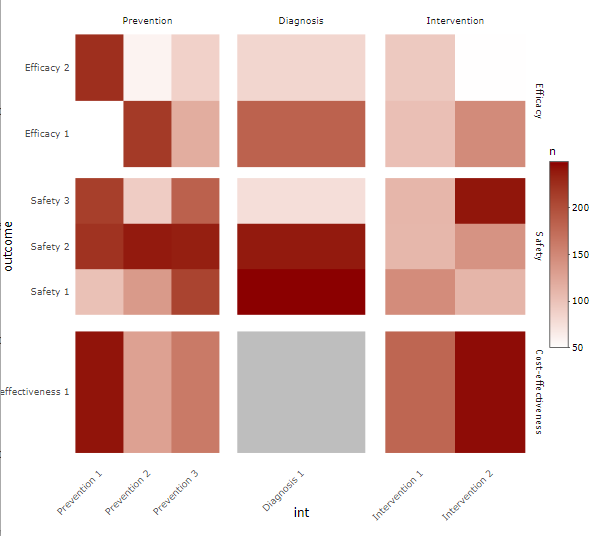
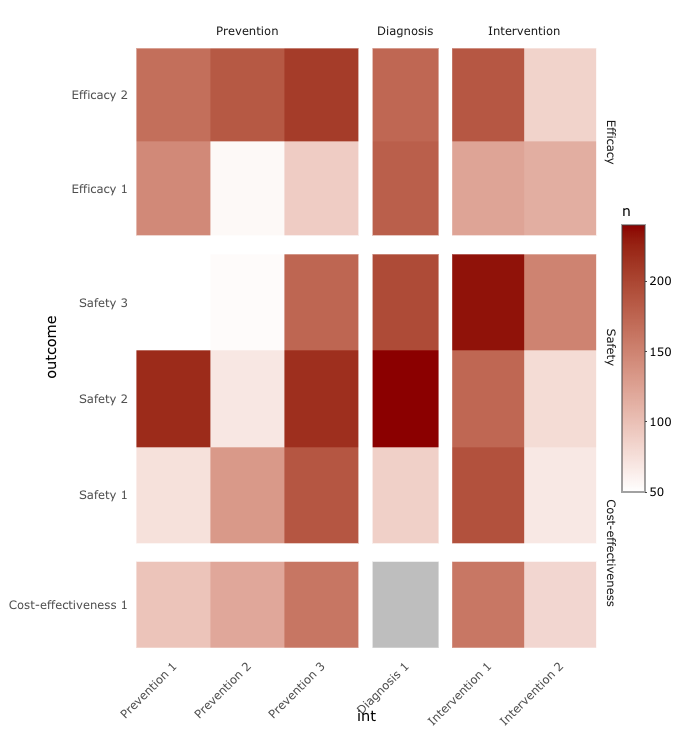
I want to make a heatmap based on the variable outcome against int, with n as the fill of each heatmap cell. Here is the desired plot:
I tried using ggplotly from the created ggplot:
plotly.df <- ggplot(df,
aes(x = int, y = outcome, fill= n))
geom_tile()
scale_fill_gradient(low="white", high="darkred")
scale_y_discrete(position = "right")
facet_grid(group.outcome ~ group.int,
scales = "free", space = "free", switch = "x")
theme_bw()
theme(axis.ticks = element_blank(),
legend.position = "left",
strip.placement = "outside",
strip.background = element_blank(),
panel.grid = element_blank(),
panel.border = element_blank(),
legend.key.height = unit(6, "lines"),
legend.title = element_blank(),
panel.spacing = unit(0, "lines"),
axis.text.x = element_text(angle = 45, vjust = 1, hjust=1))
ggplotly(plotly.df)
However, ggplotly seems to ignore space = "free" in facet_grid, so the size of the cells are not proportional:
As an alternative, I tried using subplot with plot_ly:
vals <- unique(scales::rescale(c(df$n)))
o <- order(vals, decreasing = FALSE)
cols <- scales::col_numeric("BuGn", domain = NULL)(vals)
colz <- setNames(data.frame(vals[o], cols[o]), NULL)
plot.prev <- filter(df, group.int == "Prevention") %>%
plot_ly(x = ~int, y = ~outcome, z = ~n, colorscale = colz, type = "heatmap") %>%
layout(xaxis = list(title = 'Prevention',
tickangle=315),
yaxis = list(title = "Outcome"))) %>%
hide_colorbar()
plot.diag <- filter(df, group.int == "Diagnosis") %>%
plot_ly(x = ~int, y = ~outcome, z = ~n, colorscale = colz, type = "heatmap") %>%
layout(xaxis = list(title = 'Diagnosis',
tickangle=315),
yaxis = list(ticks = "Outcome", showticklabels = FALSE)) %>%
hide_colorbar()
plot.int <- filter(df, group.int == "Intervention") %>%
plot_ly(x = ~int, y = ~outcome, z = ~n, colorscale = colz, type = "heatmap") %>%
layout(xaxis = list(title = 'Intervention',
tickangle=315),
yaxis = list(ticks = "Outcome", showticklabels = FALSE))
subplot(plot.prev, plot.diag, plot.int, widths = c(3/6, 1/6, 2/6), margin = 0)
However, I find it difficult to change axis labels and to add annotations outside the plot margin with subplot (note that I need to annotate Prevention, Diagnosis, and Treatment for the x-axis facets; and Efficacy, Safety, and Cost-effectiveness for the y-axis facets).
Is there a way to adjust facet widths with ggplotly? Or alternatively, is there an easy way to annotate subplot outside the plot margins and to change axis labels in subplot?
Thank you very much in advance
CodePudding user response:
You don't have to reinvent the wheel. Go back to the first ggplotly object. Domain is what plotly uses to govern the spaces each facet (or as it is in plotly-subplot). You can retrieve this information by assigning the ggplotly graph to an object and calling plotly_json.
However, I've worked around layout shortcuts before. You can retrieve and modify the domains like this:
p = ggplotly(plotly.df)
p$x$layout$xaxis$domain <- c(0, 1/2) # 6 blocks, 3 in this group 1/6 * 3
p$x$layout$xaxis2$domain <- c(1/2, 2/3) # start at previous position, 1 in this group
p$x$layout$xaxis3$domain <- c(2/3, 1) # remaining space
p$x$layout$yaxis3$domain <- c(0, 1/6) # 1 block in bottom chunks
p$x$layout$yaxis2$domain <- c(1/6, 2/3) # 3 in mid group
p$x$layout$yaxis$domain <- c(2/3, 1) # remaining space
p
That got me this far:
Your bottom labels are still aligned, but the top is not. Additionally, the left bottom label is cut off.
To fix the top labels I used plotly_json to figure out where they were at then used the guess-and-check method. To adjust for labels, I modified the margin.
# prevention
p$x$layout$annotations[[3]]$x <- 1/4
# diagnosis
p$x$layout$annotations[[4]]$x <- 7/12
p %>% layout(margin = list(t = 40, r = 50, b = 80, l = 130))