I'm new to R, trying to understand how plotting works.
I'm trying to plot a graph consisting of three subgraphs. I use the par(mfrow=c(1, 3)) to deal with that. But inside a loop, I want to add various lines to the three graphs. How do I choose which of the three subplots I apply the lines command to?
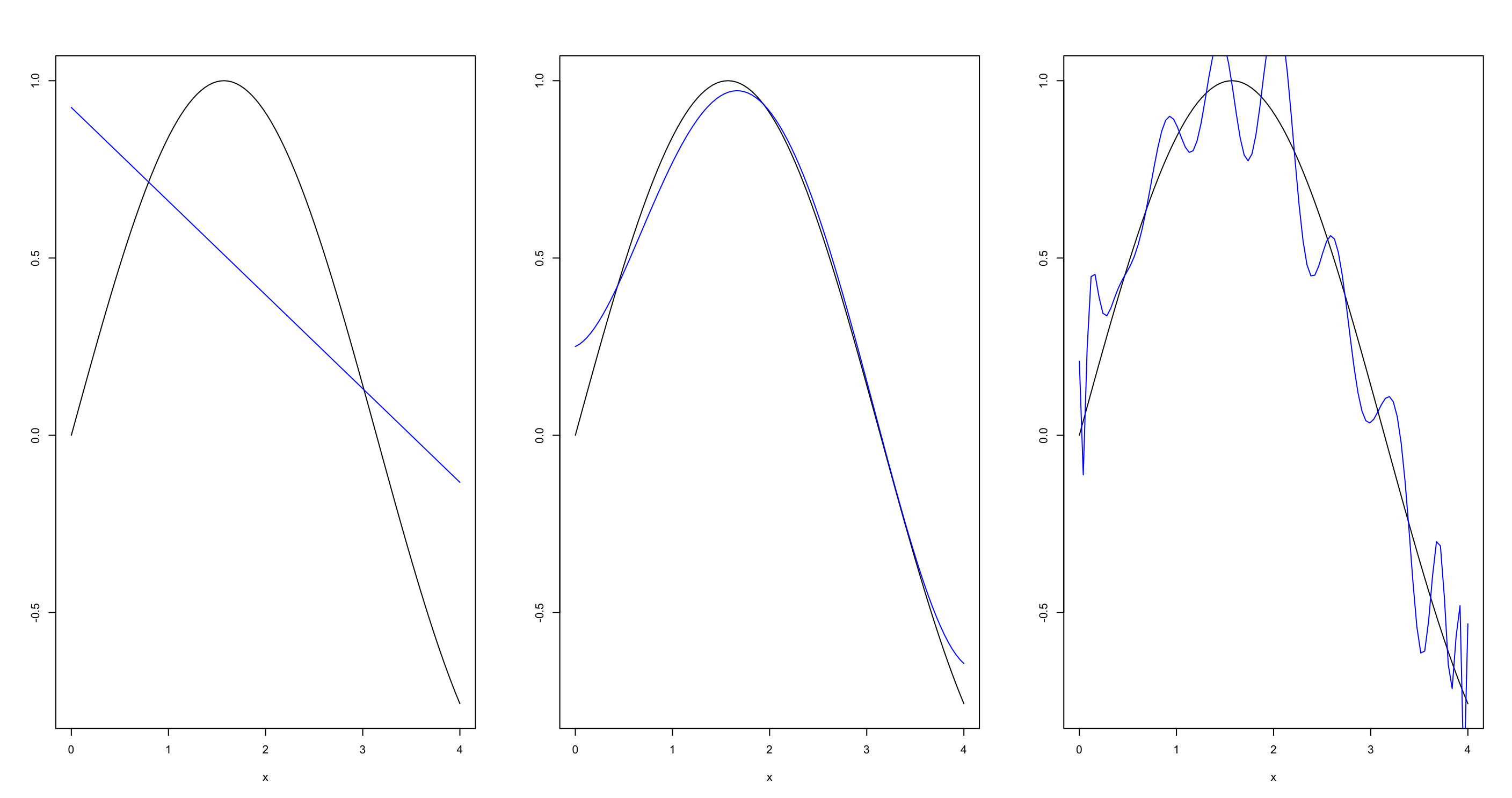
As an example, see the code below. All the lines commands get applied to the third subgraph, but this is of course not desired. See also the commands # apply lines to first figure! but how? and # apply lines to second figure! but how?
set.seed(1)
n <- 100
x <- seq(0, 4, length.out = n)
no_datasets <- 50
par(mfrow=c(1, 3))
for (i in 1:no_datasets) {
x <- seq(0, 4, length.out = n) # x <- seq(-pi, pi, length.out = n)
y <- sin(x)
errs <- rnorm(n, mean = 0, sd = 0.25) # rnorm(n) generates random numbers whose distribution is normal
t <- y errs
df <- data.frame(x, y, t, errs)
model1 <- lm(t ~ poly(x, 1), data = df)
model5 <- lm(t ~ poly(x, 5), data = df)
model25 <- lm(t ~ poly(x, 25), data = df)
if (i == 1) {
plot(df$x, df$y, xlab = "x", ylab="", col="black", type="l")
plot(df$x, df$y, xlab = "x", ylab="", col="black", type="l")
plot(df$x, df$y, xlab = "x", ylab="", col="black", type="l")
}
t_hat <- predict(model1, df)
# apply lines to first figure! but how?
lines(df$x, t_hat, col="blue")
t_hat <- predict(model5, df)
# apply lines to second figure! but how?
lines(df$x, t_hat, col="blue")
t_hat <- predict(model25, df)
# apply lines to third figure!
lines(df$x, t_hat, col="blue")
}
CodePudding user response:
Since the third plot is last, then it is just adding all the lines to the last plot. But if you nest everything in an if statement for each plot, then you will get the lines on each respective plot.
set.seed(1)
n <- 100
x <- seq(0, 4, length.out = n)
no_datasets <- 50
par(mfrow=c(1, 3))
for (i in 1:no_datasets) {
x <- seq(0, 4, length.out = n) # x <- seq(-pi, pi, length.out = n)
y <- sin(x)
errs <- rnorm(n, mean = 0, sd = 0.25) # rnorm(n) generates random numbers whose distribution is normal
t <- y errs
df <- data.frame(x, y, t, errs)
model1 <- lm(t ~ poly(x, 1), data = df)
model5 <- lm(t ~ poly(x, 5), data = df)
model25 <- lm(t ~ poly(x, 25), data = df)
if (i == 1) {
plot(df$x, df$y, xlab = "x", ylab="", col="black", type="l")
t_hat <- predict(model1, df)
lines(df$x, t_hat, col="blue")
}
if (i == 2) {
plot(df$x, df$y, xlab = "x", ylab="", col="black", type="l")
t_hat <- predict(model5, df)
lines(df$x, t_hat, col="blue")
}
if (i == 3) {
plot(df$x, df$y, xlab = "x", ylab="", col="black", type="l")
t_hat <- predict(model25, df)
lines(df$x, t_hat, col="blue")
}
}
CodePudding user response:
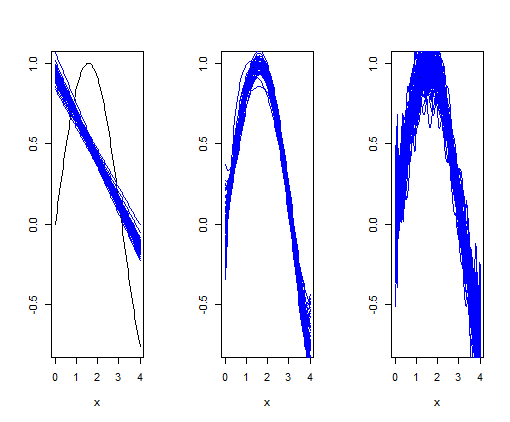
Ok, if I understand AndrewGB, it is not possible. So then the answer should be:
set.seed(1)
n <- 100
x <- seq(0, 4, length.out = n)
no_datasets <- 50
par(mfrow=c(1, 3))
polynomials <- c(1, 5, 25)
x <- seq(0, 4, length.out = n) # x <- seq(-pi, pi, length.out = n)
y <- sin(x)
for (i in 1:length(polynomials)) {
degree <- polynomials[i]
for (j in 1:no_datasets) {
errs <- rnorm(n, mean = 0, sd = 0.25) # rnorm(n) generates random numbers whose distribution is normal
t <- y errs
df <- data.frame(x, y, t, errs)
model <- lm(t ~ poly(x, degree), data = df)
if (j == 1) {
plot(df$x, df$y, xlab = "x", ylab="", col="black", type="l")
}
t_hat <- predict(model, df)
lines(df$x, t_hat, col="blue")
}
}