I am relatively new to R. I have created code to review a dataframe and identify rows of data based on specific conditions, and mark those rows with a 1 and the column "check". The code works exactly how I have intended it to with the test data. My problem is the real dataset is 1 million plus rows, and while it works, it is way too slow. I would appreciate help in improving the efficiency of this code.
#create test data
alarm <- c(0,0,0,0,0,0,1,1,0,0,0,0,0,0,0,0,1,0,0,0,0,1,0,0,0,0,0,0,0,0,0)
setpoint <- c(10,10,10,10,10,10,10,10,8,8,9,8,8,10,10,10,10,10,10,10,10,10,10,10,8,10,10,8,10,10,10)
temp <- data.frame(alarm, setpoint)
#create a new column to capture if there is any changes to setpoint after any alarm
temp$check <- ""
#review everyrow in dataframe
for(i in 1:nrow(temp)){
cat(round(i/nrow(temp)*100,2),"% \r") # prints the percentage complete in realtime.
if(temp$alarm[i]==1 && temp$setpoint[i] >= 10){
#for when alarm has occurred and the setpoint is 10 or above review the next 5 rows
for(j in 0:5){
if(temp$setpoint[i] != temp$setpoint[i j]){
#for when there has been a change in the setpoint
for(j in 0:10){
if(temp$setpoint[i] != temp$setpoint[i j]){
temp$check[i j]<-'1'
if(temp$setpoint[i j] != (temp$setpoint[i j 1])){break}
}
}
}
}
}
}
> print(temp)
alarm setpoint check
1 0 10
2 0 10
3 0 10
4 0 10
5 0 10
6 0 10
7 1 10
8 1 10
9 0 8 1
10 0 8 1
11 0 9
12 0 8
13 0 8
14 0 10
15 0 10
16 0 10
17 1 10
18 0 10
19 0 10
20 0 10
21 0 10
22 1 10
23 0 10
24 0 10
25 0 8 1
26 0 10
27 0 10
28 0 8
29 0 10
30 0 10
31 0 10
CodePudding user response:
Looking for efficiency, as the loops are already written, you could use Rcpp.
C syntax isn't very far away from R syntax, main changes are :
- declare variables
- index vectors from 0 on
- rename the third inner loop counter from
jtokbecause although keepingjworks inRit's quite error prone and not working inC - check more strictly that
i jori knever go above total number of rows
This leads to check_data.cpp:
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
DataFrame check_data(DataFrame df) {
NumericVector alarm = df["alarm"];
NumericVector setpoint = df["setpoint"];
int n = alarm.size();
LogicalVector check(n);
int i,j,k;
for(i=0; i<n;i ){
if(alarm[i]==1 && setpoint[i] >= 10){
Rcout << "pct = " << i*100/n << "%" << std::endl; // prints the percentage complete in realtime.
//for when alarm has occured and the setpoint is 10 or above review the next 5 rows
for(j=1; j<5; j ){
if (i j > n-1) break;
if(setpoint[i] != setpoint[i j]){
//for when there has been a change in the setpoint
for(k=1; k<10;k ){
if (i k> n-1) break;
if(setpoint[i] != setpoint[i k]){
check[i k] = true;
if (i k 1> n-1) break;
if(setpoint[i k] != (setpoint[i k 1])){break;}
}
}
}
}
}
}
df["check"]=check;
return(df);
}
// You can include R code blocks in C files processed with sourceCpp
// (useful for testing and development). The R code will be automatically
// run after the compilation.
//
/*** R
alarm <- c(0,0,0,0,0,0,1,1,0,0,0,0,0,0,0,0,1,0,0,0,0,1,0,0,0,0,0,0,0,0,0)
setpoint <- c(10,10,10,10,10,10,10,10,8,8,9,8,8,10,10,10,10,10,10,10,10,10,10,10,8,10,10,8,10,10,10)
temp <- data.frame(alarm, setpoint)
check_data(temp)
*/
You can then make check_data function available in your R environment by running :
library(Rcpp)
sourceCpp('check_data.cpp')
check_data(temp)
pct = 19%
pct = 22%
pct = 51%
pct = 67%
alarm setpoint check
1 0 10 FALSE
2 0 10 FALSE
3 0 10 FALSE
4 0 10 FALSE
5 0 10 FALSE
6 0 10 FALSE
7 1 10 FALSE
8 1 10 FALSE
9 0 8 TRUE
10 0 8 TRUE
11 0 9 FALSE
12 0 8 FALSE
13 0 8 FALSE
14 0 10 FALSE
15 0 10 FALSE
16 0 10 FALSE
17 1 10 FALSE
18 0 10 FALSE
19 0 10 FALSE
20 0 10 FALSE
21 0 10 FALSE
22 1 10 FALSE
23 0 10 FALSE
24 0 10 FALSE
25 0 8 TRUE
26 0 10 FALSE
27 0 10 FALSE
28 0 8 FALSE
29 0 10 FALSE
30 0 10 FALSE
31 0 10 FALSE
Performance comparison:
Unit: microseconds
expr min lq mean median uq max neval
ref 13051.8 16832.4 18510.316 18448.95 19930.10 28335.9 100
Rcpp 68.3 108.7 179.845 168.60 236.85 515.1 100
CodePudding user response:
You should investigate the logic of your algorithm. You spend a lot of time in needless looping redoing steps that were already performed (or made an error in the implementation).
I'll comment in your code
for(i in 1:nrow(temp)){
cat(round(i/nrow(temp)*100,2),"% \r") # prints the percentage complete in realtime.
printing is ok - but usually really slows things down. If you have to do consider putting it in a if (i %% 100) so it will print only every 100th line (or whatever you choose).
if(temp$alarm[i]==1 && temp$setpoint[i] >= 10){
ok - but could be vectorized (will run MUCH faster) outside the loop into temp$cond <- temp$alarm==1 & temp$setpoint >= 10
for(j in 0:5){
for both loops the condition is NEVER true for j = 0 - so start j at 1
if(temp$setpoint[i] != temp$setpoint[i j]){
for(j in 0:10){
first: don't reuse j!
Here you don't need to start j at 0 - just 2 lines above you found where it is different! you checked it. Just start at j and skip the if(temp$setpoint[i]....
if(temp$setpoint[i] != temp$setpoint[i j]){
temp$check[i j]<-'1'
if(temp$setpoint[i j] != (temp$setpoint[i j 1])){break}
}
}
break HERE! otherwise the inner loop might rerun times doing the exactly same!
}
}
}
}
CodePudding user response:
Edit:
This answer provides the correct answer with the example dataset but not with @Luke_DataSci's actual dataset.
Original answer:
Here is a potential 'brute force' solution that should be significantly faster:
library(dplyr)
alarm <- c(0,0,0,0,0,0,1,1,0,0,0,0,0,0,0,0,1,0,0,0,0,1,0,0,0,0,0,0,0,0,0)
setpoint <- c(10,10,10,10,10,10,10,10,8,8,8,8,8,10,10,10,10,10,10,10,10,10,10,10,8,10,10,8,10,10,10)
test_dataset_1 <- data.frame(alarm, setpoint)
alarm2 <- c(0,0,0,0,0,0,1,1,0,0,0,0,0,0,0,0,1,0,0,0,0,1,0,0,0,0,0,0,0,0,0)
setpoint2 <- c(10,10,10,10,10,10,10,10,8,8,9,8,8,10,10,10,10,10,10,10,10,10,10,10,8,10,10,8,10,10,10)
test_dataset_2 <- data.frame(alarm2, setpoint2)
ifelse_func <- function(df){
df$check <- ifelse(
(lag(df$alarm, n = 1, default = 0) == 1 &
lag(df$setpoint, n = 1, default = 0) >= 10 &
df$setpoint != 10) |
(lag(df$alarm, n = 2, default = 0) == 1 &
lag(df$setpoint, n = 2, default = 0) >= 10 &
df$setpoint != 10 &
df$setpoint == lag(df$setpoint, n = 1, default = 0)) |
(lag(df$alarm, n = 3, default = 0) == 1 &
lag(df$setpoint, n = 3, default = 0) >= 10 &
df$setpoint != 10 &
(df$setpoint == lag(df$setpoint, n = 1, default = 0) |
lag(df$setpoint, n = 1, default = 0) == 10) &
(df$setpoint == lag(df$setpoint, n = 2, default = 0) |
lag(df$setpoint, n = 2, default = 0) == 10)) |
(lag(df$alarm, n = 4, default = 0) == 1 &
lag(df$setpoint, n = 4, default = 0) >= 10 &
df$setpoint != 10 &
(df$setpoint == lag(df$setpoint, n = 1, default = 0) |
lag(df$setpoint, n = 1, default = 0) == 10) &
(df$setpoint == lag(df$setpoint, n = 2, default = 0) |
lag(df$setpoint, n = 2, default = 0) == 10) &
(df$setpoint == lag(df$setpoint, n = 3, default = 0) |
lag(df$setpoint, n = 3, default = 0) == 10)) |
(lag(df$alarm, n = 5, default = 0) == 1 &
lag(df$setpoint, n = 5, default = 0) >= 10 &
df$setpoint != 10 &
(df$setpoint == lag(df$setpoint, n = 1, default = 0) |
lag(df$setpoint, n = 1, default = 0) == 10) &
(df$setpoint == lag(df$setpoint, n = 2, default = 0) |
lag(df$setpoint, n = 2, default = 0) == 10) &
(df$setpoint == lag(df$setpoint, n = 3, default = 0) |
lag(df$setpoint, n = 3, default = 0) == 10) &
(df$setpoint == lag(df$setpoint, n = 4, default = 0) |
lag(df$setpoint, n = 4, default = 0) == 10)),
1, "")
return(df)
}
forloop_func <- function(df){
df$check <- ""
for(i in 1:nrow(df)){
# cat(round(i/nrow(temp)*100,2),"% \r") # prints the percentage complete in realtime.
if(df$alarm[i]==1 && df$setpoint[i] >= 10){
#for when alarm has occurred and the setpoint is 10 or above review the next 5 rows
for(j in 0:5){
if(df$setpoint[i] != df$setpoint[i j]){
#for when there has been a change in the setpoint
for(j in 0:10){
if(df$setpoint[i] != df$setpoint[i j]){
df$check[i j]<-'1'
if(df$setpoint[i j] != (df$setpoint[i j 1])){break}
}
}
}
}
}
}
return(df)
}
all_equal(ifelse_func(test_dataset_1), forloop_func(test_dataset_1))
#> [1] TRUE
all_equal(ifelse_func(test_dataset_2), forloop_func(test_dataset_2))
#> [1] TRUE
library(microbenchmark)
library(ggplot2)
res <- microbenchmark(ifelse_func(test_dataset_2),
forloop_func(test_dataset_2),
times = 10)
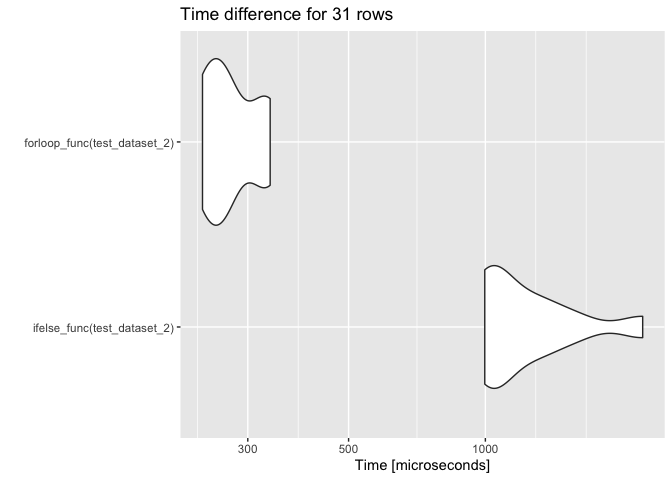
autoplot(res) ggtitle("Time difference for 31 rows")
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
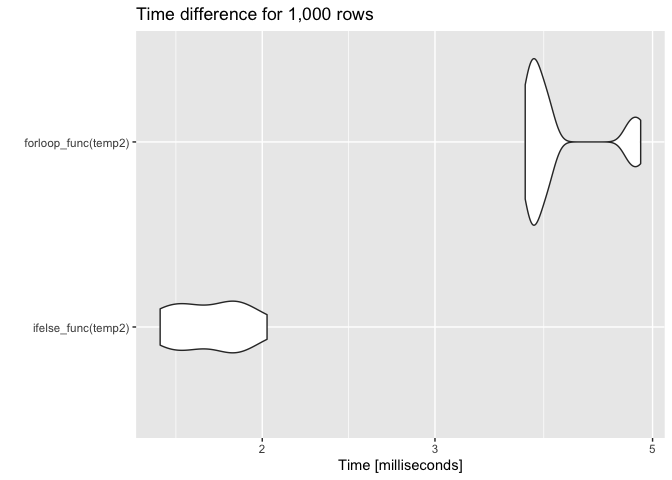
set.seed(123)
temp2 <- data.frame(alarm = sample(alarm, 1000, replace = TRUE),
setpoint = sample(setpoint, 1000, replace = TRUE))
res2 <- microbenchmark(ifelse_func(temp2), forloop_func(temp2), times = 10)
autoplot(res2) ggtitle("Time difference for 1,000 rows")
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
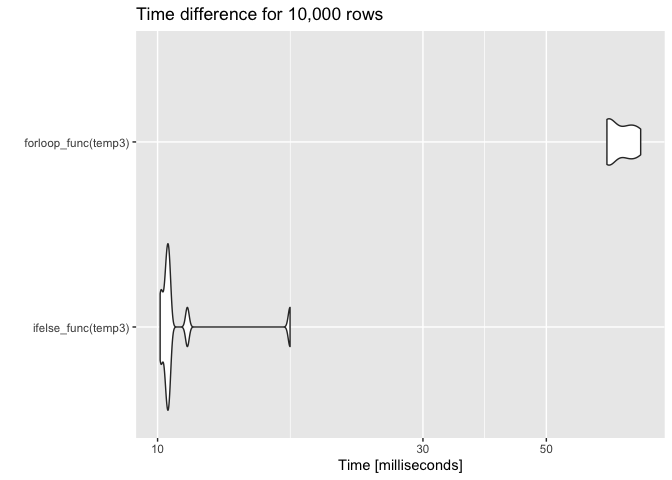
temp3 <- data.frame(alarm = sample(alarm, 10000, replace = TRUE),
setpoint = sample(setpoint, 10000, replace = TRUE))
res3 <- microbenchmark(ifelse_func(temp3), forloop_func(temp3), times = 10)
autoplot(res3) ggtitle("Time difference for 10,000 rows")
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
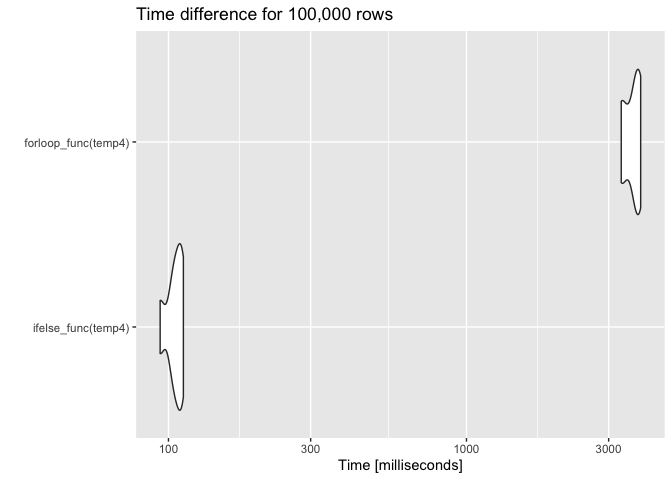
temp4 <- data.frame(alarm = sample(alarm, 100000, replace = TRUE),
setpoint = sample(setpoint, 100000, replace = TRUE))
res4 <- microbenchmark(ifelse_func(temp4), forloop_func(temp4), times = 6)
autoplot(res4) ggtitle("Time difference for 100,000 rows")
#> Coordinate system already present. Adding new coordinate system, which will replace the existing one.
For 1 millions rows:
temp5 <- data.frame(alarm = sample(alarm, 1000000, replace = TRUE),
setpoint = sample(setpoint, 1000000, replace = TRUE))
Unit: milliseconds
expr min lq mean median uq max neval cld
ifelse_func(temp5) 873.8556 873.8556 1181.997 1181.997 1490.138 1490.138 2 a
forloop_func(temp5) 292242.7181 292242.7181 295101.463 295101.463 297960.208 297960.208 2 b
Created on 2022-04-07 by the reprex package (v2.0.1)
So, despite being ~3X slower than your for-loop method with 31 rows, this approach is ~250X faster with 1 million rows.
Now the question is whether or not it provides the correct answer...
CodePudding user response:
If I understand your objective properly, perhaps you can try the data.table approach like below
library(data.table)
setDT(temp)[
,
check := ({
d <- cumsum(c(FALSE, diff(setpoint)) != 0) == 1
d & min(c(which(d), Inf)) <= 5
}),
cumsum(alarm == 1 & setpoint >= 10)
]
which gives
alarm setpoint check
1: 0 10 0
2: 0 10 0
3: 0 10 0
4: 0 10 0
5: 0 10 0
6: 0 10 0
7: 1 10 0
8: 1 10 0
9: 0 8 1
10: 0 8 1
11: 0 9 0
12: 0 8 0
13: 0 8 0
14: 0 10 0
15: 0 10 0
16: 0 10 0
17: 1 10 0
18: 0 10 0
19: 0 10 0
20: 0 10 0
21: 0 10 0
22: 1 10 0
23: 0 10 0
24: 0 10 0
25: 0 8 1
26: 0 10 0
27: 0 10 0
28: 0 8 0
29: 0 10 0
30: 0 10 0
31: 0 10 0
