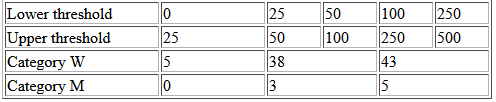
I would like to automatically create a table in R Markdown, where the table is created in html, but the data comes from R. Could anyone help me with how to do this?
R data:
dat <- structure(list(rn = c("W", "M"), `[ 0, 25)` = c(5L,
0L), `[ 25, 50)` = c(0L, 0L), `[ 25, 100)` = c(38L,
3L), `[ 50, 100)` = c(0L, 0L), `[ 100, 250)` = c(43L,
5L), `[ 100, 500)` = c(0L, 0L)), row.names = c(NA, -2L), class = c("data.table",
"data.frame"))
rn [ 0, 25) [ 25, 50) [ 25, 100) [ 50, 100) [ 100, 250) [ 100, 500)
1: W 5 0 38 0 43 0
2: M 0 0 3 0 5 0
Html code:
<html>
<table border="1">
<tr>
<td width="150">Lower threshold</td>
<td width="50">0</td>
<td width="50">25</td>
<td width="50">50</td>
<td width="50">100</td>
<td width="50">250</td>
<tr>
<td width="150">Upper threshold</td>
<td width="50">25</td>
<td width="50">50</td>
<td width="50">100</td>
<td width="50">250</td>
<td width="50">500</td>
<tr>
<td width="150">Category W</td>
<td width="100" colspan="1">5</td>
<td width="100" colspan="2">38</td>
<td width="100" colspan="2">5</td>
</tr>
<tr>
<td width="150">Category M</td>
<td width="100" colspan="1">0</td>
<td width="100" colspan="2">3</td>
<td width="100" colspan="2">5</td>
</tr>
CodePudding user response:
One approach to create your HTML table from scratch which makes heavy use of htmltools and purrr may look like so. To make this work you have to do some data wrangling to put your data and the info needed to create your data in a tidy data format:
---
title: "HTML Table"
date: '2022-04-19'
output: html_document
---
```{r echo=FALSE}
dat <- structure(list(
row = structure(c(
1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L, 3L, 3L, 3L, 4L, 4L, 4L
), .Label = c(
"Lower threshold",
"Upper threshold", "Category W", "Category M"
), class = "factor"),
value = c(
0, 25, 50, 100, 250, 25, 50, 100, 250, 500, 5,
38, 5, 0, 3, 5
), width = c(
50, 50, 50, 50, 50, 50, 50, 50,
50, 50, 100, 100, 100, 100, 100, 100
), colspan = c(
NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, 1, 2, 2, 1, 2, 2
)
), row.names = c(
NA,
-16L
), class = "data.frame")
```
```{r}
head(dat)
```
```{r echo=FALSE}
make_html_table_row <- function(row, label) {
htmltools::tagList(
htmltools::tags$td(label, width = "150"),
purrr::pmap(row, function(row, value, width, colspan) {
htmltools::tags$td(value, width = width, colspan = if (!is.na(colspan)) colspan)
})
) |>
htmltools::tags$tr()
}
make_html_table <- function(x) {
x_split <- split(x, x$row)
purrr::imap(x_split, make_html_table_row) |>
htmltools::tagList() |>
htmltools::tags$table(border = "1") |>
htmltools::tags$html()
}
```
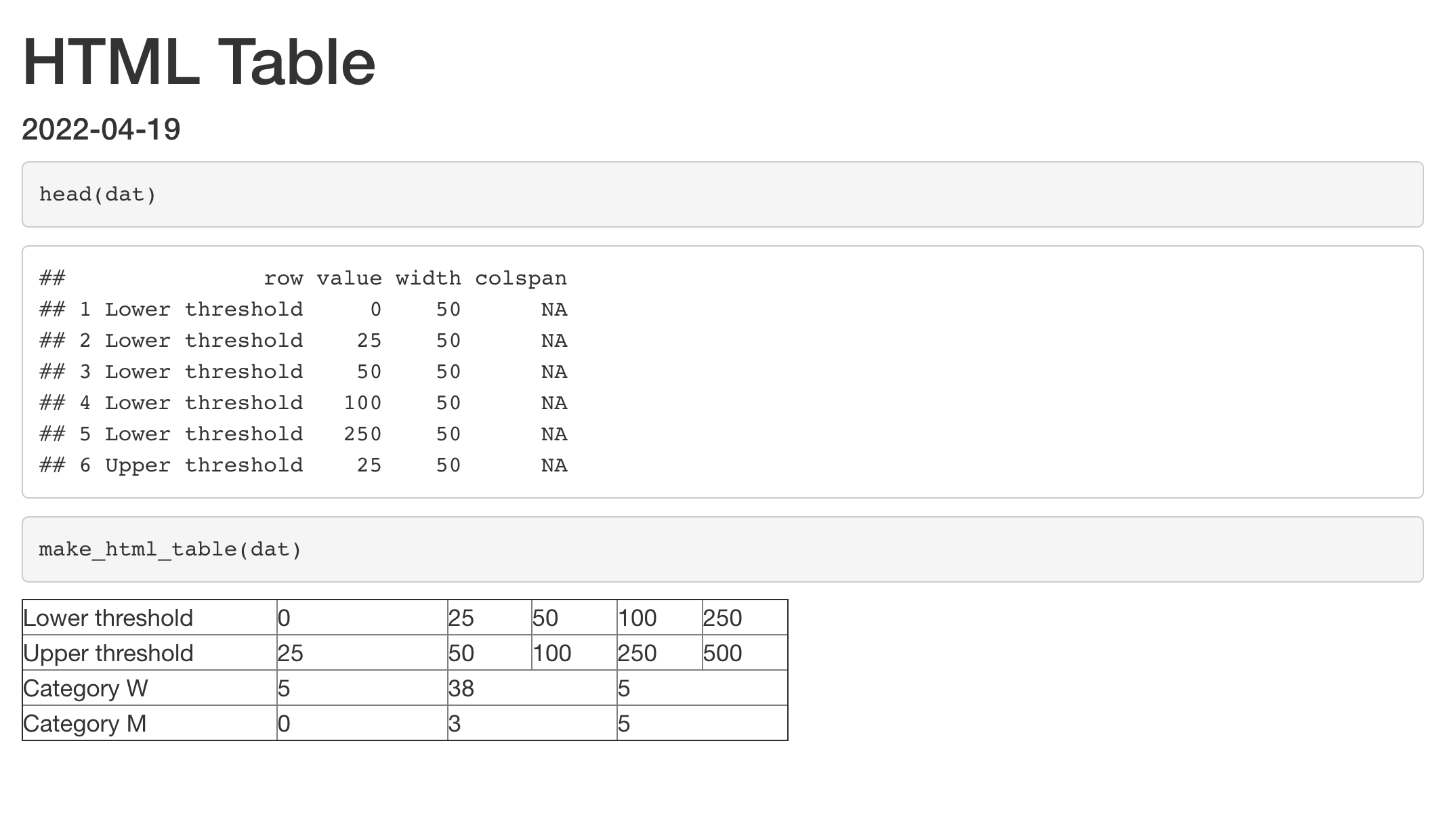
```{r results='asis'}
make_html_table(dat)
```
DATA
Code used to create the data.
dat <- list(
data.frame(
row = "Lower threshold",
value = c(0, 25, 50, 100, 250),
width = 50,
colspan = NA
),
data.frame(
row = "Upper threshold",
value = c(25, 50, 100, 250, 500),
width = 50,
colspan = NA
),
data.frame(
row = "Category W",
value = c(5, 38, 5),
width = 100,
colspan = c(1, 2, 2)
),
data.frame(
row = "Category M",
value = c(0, 3, 5),
width = 100,
colspan = c(1, 2, 2)
)
) |>
dplyr::bind_rows()
dat$row <- factor(dat$row, levels = c("Lower threshold", "Upper threshold", "Category W", "Category M"))