I am trying to create a fixation maps where the weight of each fixation on the 2d density map is determined by its duration. As I understood, the stat_density2d() function accepts the weight argument but does not process it (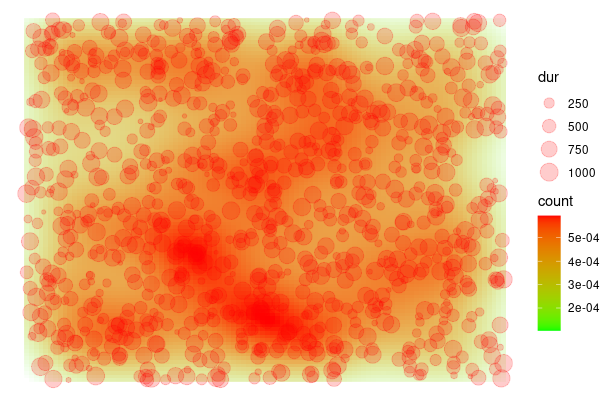
CodePudding user response:
Not a ggplot2 user, but basically you want to estimate a weighted 2d-density and make an image out of it. Your 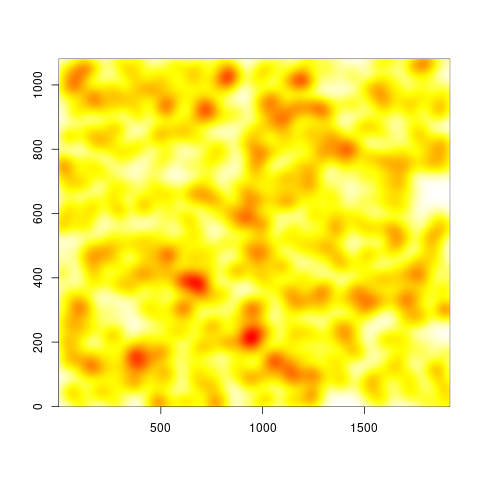
This almost runs forever, though with n=1000...
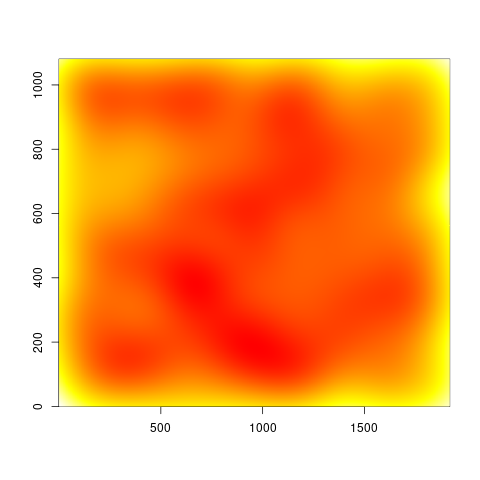
Weighted 2d density estimation
In a comment on the answer mentioned above, 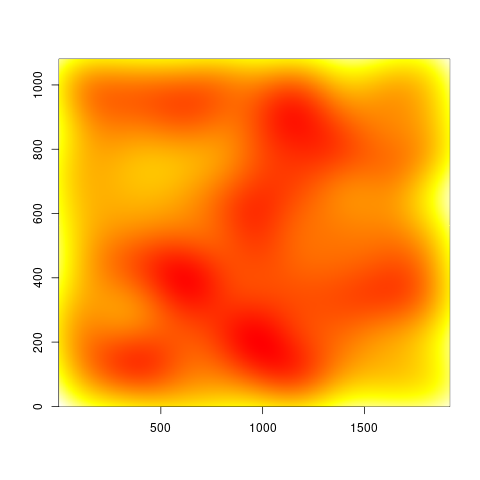
However, the two versions look quite different, and I can't tell which is right, since I am not really an expert with this method. But at least there is a noticeable difference to the unweighted image:
Unweighted image
dens0 <- MASS::kde2d(df$x, df$y, n=n)
image(dens0, col=heat.colors(n, rev=TRUE))
Points
Still adding the points might be pointless, but you could run this line after image:
points(y ~ x, df, cex=proportions(dur)*2e3, col='green')
Taken from r-help (Ort 2006):
kde2d.weighted <- function(x, y, w, h, n=n, lims=c(range(x), range(y))) {
nx <- length(x)
if (length(y) != nx)
stop("data vectors must be the same length")
gx <- seq(lims[1], lims[2], length=n) ## gridpoints x
gy <- seq(lims[3], lims[4], length=n) ## gridpoints y
if (missing(h))
h <- c(MASS::bandwidth.nrd(x), MASS::bandwidth.nrd(y))
if (missing(w))
w <- numeric(nx) 1
h <- h/4
ax <- outer(gx, x, "-")/h[1] ## distance of each point to each grid point in x-direction
ay <- outer(gy, y, "-")/h[2] ## distance of each point to each grid point in y-direction
z <- (matrix(rep(w,n), nrow=n, ncol=nx, byrow=TRUE)*
matrix(dnorm(ax), n, nx)) %*%
t(matrix(dnorm(ay), n, nx))/(sum(w)*h[1]*h[2]) ## z is the density
return(list(x=gx, y=gy, z=z))
}
CodePudding user response:
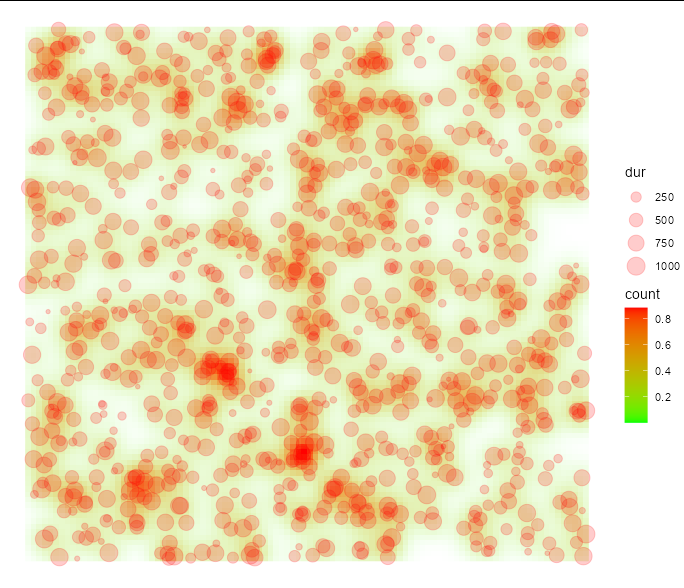
The simplest way round this is to use tidyr::uncount to replicate the rows of your data frame, using dur as weights:
library(ggplot2)
ggplot(tidyr::uncount(df, dur), aes(x=x, y =y))
stat_density2d(geom='raster',
aes(fill=..count.., alpha=..count..), contour=FALSE)
geom_point(data = df, aes(size=dur), alpha=0.2, color="red")
scale_fill_gradient(low="green", high="red")
scale_alpha_continuous(range=c(0, 1) , guide="none")
theme_void()
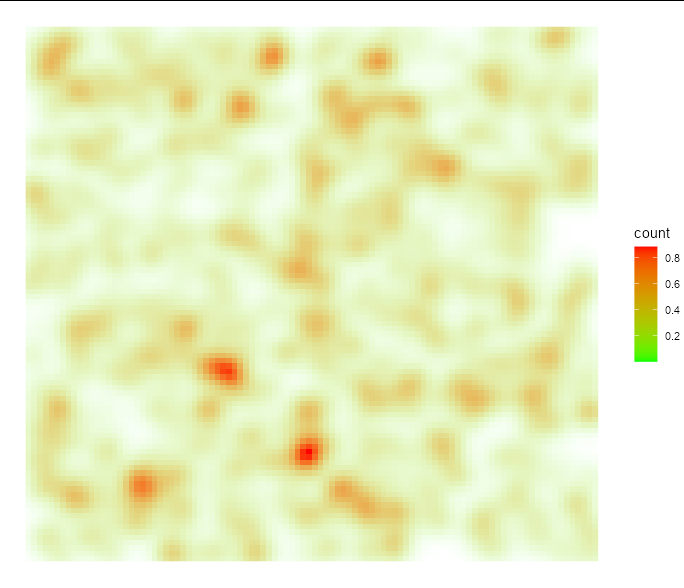
The effect is probably easier to see with the points removed:
ggplot(tidyr::uncount(df, dur), aes(x=x, y =y))
stat_density2d(geom='raster',
aes(fill=..count.., alpha=..count..), contour=FALSE)
scale_fill_gradient(low="green", high="red")
scale_alpha_continuous(range=c(0, 1) , guide="none")
theme_void()