I have a dataframe with multiple columns, and I need to do a fishers test for each column. I know that I first need to change each column into a 2x2 contingency table and then do a fishers test on it and then loop through the entire dataframe (containing 1000 columns).
For instance, the dataframe will look like this, and I think I should convert it to this form inorder to do the fishers test.
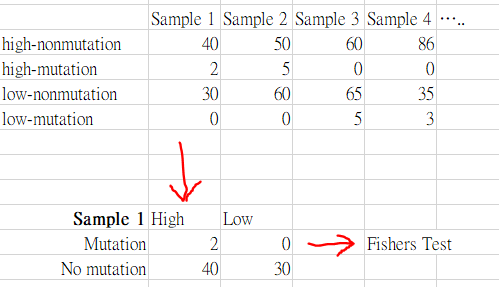
Is there any way to do this? Is there a way to do the fishers test without having to convert it into a contingency table? I'm stuck on how to do fishers test for this dataframe..thank you very much!
CodePudding user response:
First provide the data in reproducible format with dput():
mydata <- structure(list(Sample1 = c(40, 2, 30, 0), Sample2 = c(50, 5,
60, 0), Sample3 = c(60, 0, 65, 5), Sample4 = c(86, 0, 35, 3)), class = "data.frame", row.names = c("high-nonmutation",
"high-mutation", "low-nonmutation", "low-mutation"))
Now re-arrange the rows so they match your table:
mydata <- mydata[c(2, 1, 4, 3), ]
Finally use apply() to run the Fisher's tests:
results <- apply(mydata, 2, function(x) fisher.test(matrix(x, 2, 2)))
The object results is a list. Each list part is a test:
results[[1]] # or results[["Sample1"]]
#
# Fisher's Exact Test for Count Data
#
# data: matrix(x, 2, 2)
# p-value = 0.507
# alternative hypothesis: true odds ratio is not equal to 1
# 95 percent confidence interval:
# 0.1341153 Inf
# sample estimates:
# odds ratio
# Inf
CodePudding user response:
This answer uses the data as posted in dcarlson's answer.
First see how to run one test, for instance, for Sample1.
mydata <- structure(list(
Sample1 = c(40, 2, 30, 0),
Sample2 = c(50, 5, 60, 0),
Sample3 = c(60, 0, 65, 5),
Sample4 = c(86, 0, 35, 3)),
class = "data.frame",
row.names = c("high-nonmutation", "high-mutation",
"low-nonmutation", "low-mutation"))
suppressPackageStartupMessages({
library(dplyr)
library(tidyr)
})
mydata %>%
tibble::rownames_to_column("rownms") %>%
separate(rownms, into = c("Rank", "Mutation")) %>%
xtabs(Sample1 ~ Mutation Rank, data = .) %>%
fisher.test()
#>
#> Fisher's Exact Test for Count Data
#>
#> data: .
#> p-value = 0.507
#> alternative hypothesis: true odds ratio is not equal to 1
#> 95 percent confidence interval:
#> 0.1341153 Inf
#> sample estimates:
#> odds ratio
#> Inf
Created on 2022-04-23 by the reprex package (v2.0.1)
Now run all test with broom package function tidy.
mydata <- structure(list(
Sample1 = c(40, 2, 30, 0),
Sample2 = c(50, 5, 60, 0),
Sample3 = c(60, 0, 65, 5),
Sample4 = c(86, 0, 35, 3)),
class = "data.frame",
row.names = c("high-nonmutation", "high-mutation",
"low-nonmutation", "low-mutation"))
suppressPackageStartupMessages({
library(dplyr)
library(tidyr)
library(broom)
})
mydata %>%
tibble::rownames_to_column("rownms") %>%
separate(rownms, into = c("Rank", "Mutation")) %>%
pivot_longer(cols = starts_with("Sample"), names_to = "Sample") %>%
group_by(Sample) %>%
do(tidy(fisher.test( xtabs(value ~ Mutation Rank, data = .data) ), data = .x))
#> # A tibble: 4 x 7
#> # Groups: Sample [4]
#> Sample estimate p.value conf.low conf.high method alternative
#> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 Sample1 Inf 0.507 0.134 Inf Fisher's Exact Test f~ two.sided
#> 2 Sample2 Inf 0.0227 1.04 Inf Fisher's Exact Test f~ two.sided
#> 3 Sample3 0 0.0614 0 1.24 Fisher's Exact Test f~ two.sided
#> 4 Sample4 0 0.0272 0 1.04 Fisher's Exact Test f~ two.sided
Created on 2022-04-23 by the reprex package (v2.0.1)
