I have a function which returns a multidimensional array of k clusters. My algorith works for the most part, but I need it to return a categorical array instead of a multidimensional array. Here is my code:
import numpy as np
import pandas as pd
import random
from bokeh.sampledata.iris import flowers
from typing import List, Tuple
def get_closest(data_point: np.ndarray, centroids: np.ndarray):
"""
Takes a data_point and a nd.array of multiple centroids and returns the index of the centroid closest to data_point
by computing the euclidean distance for each centroid and picking the closest.
"""
N = centroids.shape[0]
dist = np.empty(N)
for i, c in enumerate(centroids):
dist[i] = np.linalg.norm(c - data_point)
index_min = np.argmin(dist)
return index_min
# Use these centroids in the first iteration of you algorithm if "Random Centroids" is set to False in the Dashboard
DEFAULT_CENTROIDS = np.array([[5.664705882352942, 3.0352941176470587, 3.3352941176470585, 1.0176470588235293],
[5.446153846153847, 3.2538461538461543, 2.9538461538461536, 0.8846153846153846],
[5.906666666666667, 2.933333333333333, 4.1000000000000005, 1.3866666666666667],
[5.992307692307692, 3.0230769230769234, 4.076923076923077, 1.3461538461538463],
[5.747619047619048, 3.0714285714285716, 3.6238095238095243, 1.1380952380952383],
[6.161538461538462, 3.030769230769231, 4.484615384615385, 1.5307692307692309],
[6.294117647058823, 2.9764705882352938, 4.494117647058823, 1.4],
[5.853846153846154, 3.215384615384615, 3.730769230769231, 1.2076923076923078],
[5.52857142857143, 3.142857142857143, 3.107142857142857, 1.007142857142857],
[5.828571428571429, 2.9357142857142855, 3.664285714285714, 1.1]])
def k_means(data_np: np.ndarray, k:int=3, n_iter:int=500, random_initialization=False) -> Tuple[np.ndarray, int]:
"""
:param data: your data, a numpy array with shape (n_entries, n_features)
:param k: The number of clusters to compute
:param n_iter: The maximal numnber of iterations
:param random_initialization: If False, DEFAULT_CENTROIDS are used as the centroids of the first iteration.
:return: A tuple (cluster_indices: A numpy array of cluster_indices,
n_iterations: the number of iterations it took until the algorithm terminated)
"""
# Initialize the algorithm by assigning random cluster labels to each entry in your dataset
k=k 1
centroids = data_np[random.sample(range(len(data_np)), k)]
labels = np.array([np.argmin([(el - c) ** 2 for c in centroids]) for el in data_np])
clustering = []
for k in range(k):
clustering.append(data_np[labels == k])
# Implement K-Means with a while loop, which terminates either if the centroids don't move anymore, or
# if the number of iterations exceeds n_iter
counter = 0
while counter < n_iter:
# Compute the new centroids, if random_initialization is false use DEFAULT_CENTROIDS in the first iteration
# if you use DEFAULT_CENTROIDS, make sure to only pick the k first entries from them.
if random_initialization is False and counter == 0:
centroids = DEFAULT_CENTROIDS[random.sample(range(len(DEFAULT_CENTROIDS)), k)]
# Update the cluster labels using get_closest
labels = np.array([get_closest(el, centroids) for el in data_np])
clustering = []
for i in range(k):
clustering.append(np.where(labels == i)[0])
counter = 1
new_centroids = np.zeros_like(centroids)
for i in range(k):
if len(clustering[i]) > 0:
new_centroids[i] = data_np[clustering[i]].mean(axis=0)
else:
new_centroids[i] = centroids[i]
# if the centroids didn't move, exit the while loop
if clustering is not None and (centroids == new_centroids).sum() == 0:
break
else:
centroids = new_centroids
pass
# return the final cluster labels and the number of iterations it took
return clustering, counter
# read and store the dataset
data: pd.DataFrame = flowers.copy(deep=True)
data = data.drop(['species'], axis=1)
data_np = np.asarray(data)
clustering, counter = k_means(data_np,4,500,False)
So clustering looks like so
clustering
[array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33,
34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 57,
98], dtype=int64),
array([60, 93], dtype=int64),
array([ 50, 51, 52, 53, 54, 55, 56, 58, 61, 62, 63, 65, 66,
67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79,
80, 81, 82, 83, 86, 87, 89, 90, 91, 92, 94, 95, 96,
97, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110,
111, 112, 113, 114, 115, 116, 117, 118, 119, 120, 121, 122, 123,
124, 125, 126, 127, 128, 129, 130, 131, 132, 133, 134, 135, 136,
137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147, 148, 149],
dtype=int64),
array([59, 64, 84, 85, 88], dtype=int64)]
However, what I'm looking for is an array like
clustering
array([1, 3, 2, ..., 4, 1, 4], dtype=int64)]
Also, the while loop is always terminating after 1 iteration which shouldn't be the case.
counter
1
EDIT1: The code continues as follows.
def callback(attr, old, new):
# recompute the clustering and update the colors of the data points based on the result
k = slider_k.valued_throttled
init = select_init.value
clustering_new, counter_new = k_means(data_np,k,500,init)
pass
# Create the dashboard
# 1. A Select widget to choose between random initialization or using the DEFAULT_CENTROIDS on top
select_init = Select(title='Random Centroids',value='False',options=['True','False'])
# 2. A Slider to choose a k between 2 and 10 (k being the number of clusters)
slider_k = Slider(start=2,end=10,value=3,step=1,title='k')
# 4. Connect both widgets to the callback
select_init.on_change('value',callback)
slider_k.on_change('value_throttled',callback)
# 3. A ColumnDataSource to hold the data and the color of each point you need
source = ColumnDataSource(dict(petal_length=data['petal_length'],sepal_length=data['sepal_length'],petal_width=data['petal_width'],clustering=clustering))
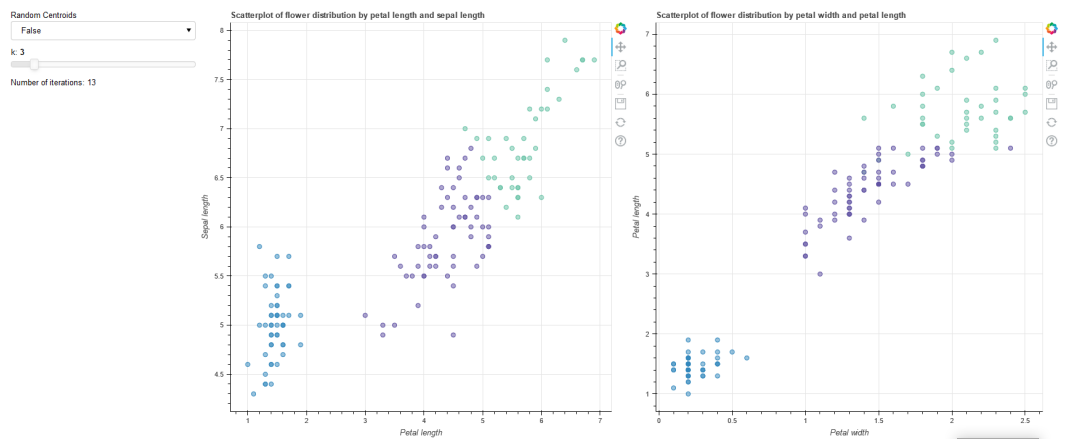
# 4. Two plots displaying the dataset based on the following table, have a look at the images
# in the handout if this confuses you.
#
# Axis/Plot Plot1 Plot2
# X Petal length Petal width
# Y Sepal length Petal length
#
# Use a categorical color mapping, such as Spectral10, have a look at this section of the bokeh docs:
# https://docs.bokeh.org/en/latest/docs/user_guide/categorical.html#filling
plot1 = figure(plot_width=100,plot_height=100,title='Scatterplot of flowers distribution by petal length and sepal length')
plot1.yaxis.axis_label = 'Sepal length'
plot1.xaxis.axis_label = 'Petal length'
scatter1 = plot1.scatter(x='petal_length',y='sepal_length',source=source,fill_color=factor_cmap('clustering', palette=Spectral10, factors=clustering))
plot2 = figure(plot_width=100,plot_height=100,title='Scatterplot of flowers distribution by petal width and petal length')
plot2.yaxis.axis_label = 'Petal length'
plot2.xaxis.axis_label = 'Petal width'
scatter2 = plot2.scatter(x='petal_width',y='petal_length',source=source,fill_color=factor_cmap('clustering', palette=Spectral10, factors=clustering))
# 5. A Div displaying the currently number of iterations it took the algorithm to update the plot.
div = Div(text='Number of iterations: ')
Thus the end result should look like so
CodePudding user response:
I'm not sure I understand what you need.
If clustering contains a list of arrays where each array represent a cluster and the ith array contains the indices of the samples that belong to the ith cluster and what you need is to convert this to a single vector of size number_of_samples that represent the cluster each sample belongs to you can do it like this:
def to_classes(clustering):
# Get number of samples (you can pass it directly to the function)
num_samples = sum(x.shape[0] for x in clustering)
indices = np.empty((num_samples,)) # An empty array with correct size
for ith, cluster in enumerate(clustering):
# use cluster indices to assign to correct the cluster index
indices[cluster] = ith
return indices
The loops exists after a single iteration because the break condition is wrong, I think what you want is actually
# note the !=
if clustering is not None and (centroids != new_centroids).sum() == 0:
break
