Suppose we start with the below data dataframe, generated by the code immediately beneath:
> data
ID Period_1 Period_2 Values State
1 1 1 2020-01 5 X0
2 1 2 2020-02 10 X1
3 1 3 2020-03 15 X0
4 2 1 2020-04 0 X0
5 2 2 2020-05 2 X2
6 2 3 2020-06 4 X0
7 3 1 2020-02 3 X2
8 3 2 2020-03 6 X1
9 3 3 2020-04 9 X0
data <-
data.frame(
ID = c(1,1,1,2,2,2,3,3,3),
Period_1 = c(1, 2, 3, 1, 2, 3, 1, 2, 3),
Period_2 = c("2020-01","2020-02","2020-03","2020-04","2020-05","2020-06","2020-02","2020-03","2020-04"),
Values = c(5, 10, 15, 0, 2, 4, 3, 6, 9),
State = c("X0","X1","X0","X0","X2","X0", "X2","X1","X0")
)
I am trying to learn how to use R package data.table, and would like to use it to count transitions out of a given state (state "X0" in the below code example) to another state, when moving or "transitioning" from one period to the next (in this case, period measurement is "Period_1"). I get the following results when running the data.table code immediately beneath:
OutflowState 2 4
1: X0 0 0
2: X1 1 0
3: X2 1 0
Code run:
library(data.table)
dcast(
setDT(data)[, OutflowState := factor(shift(State, type = c("lead"))), by = ID]
[, period_factor := lapply(.SD, factor), .SDcols = "Period_1"]
[, period_factor := as.numeric(period_factor) 1],
OutflowState ~ period_factor, length,
value.var = "Values", subset = .(State == "X0"), drop = FALSE
)
This output is correct, but I would like to (a) add columns to the output for periods 1 and 3 (period 1 will always be all 0's and period 3 should show all 0's in the case of this data dataframe because there were no state = X0 in periods 2; and (b) drop from the output the column where Period_1 = 4, because there is no period = 4, it's just a trick used in the code above as.numeric(period_factor) 1 in order to flag the next transition period. How could I do this?
I get the following interim dataframe when running the segment of code shown beneath it, so one solution is to knock out any rows where OutflowState = NA (eliminating all notional period 4's) but I don´t know how to do this.
ID Period_1 Period_2 Values State OutflowState period_factor
1: 1 1 2020-01 5 X0 X1 2
2: 1 2 2020-02 10 X1 X0 3
3: 1 3 2020-03 15 X0 <NA> 4
4: 2 1 2020-04 0 X0 X2 2
5: 2 2 2020-05 2 X2 X0 3
6: 2 3 2020-06 4 X0 <NA> 4
7: 3 1 2020-02 3 X2 X1 2
8: 3 2 2020-03 6 X1 X0 3
9: 3 3 2020-04 9 X0 <NA> 4
setDT(data)[, OutflowState := factor(shift(State, type = c("lead"))), by = ID][
, period_factor := lapply(.SD, factor), .SDcols = "Period_1"][
, period_factor := as.numeric(period_factor) 1
]
data
This question is an outgrowth of 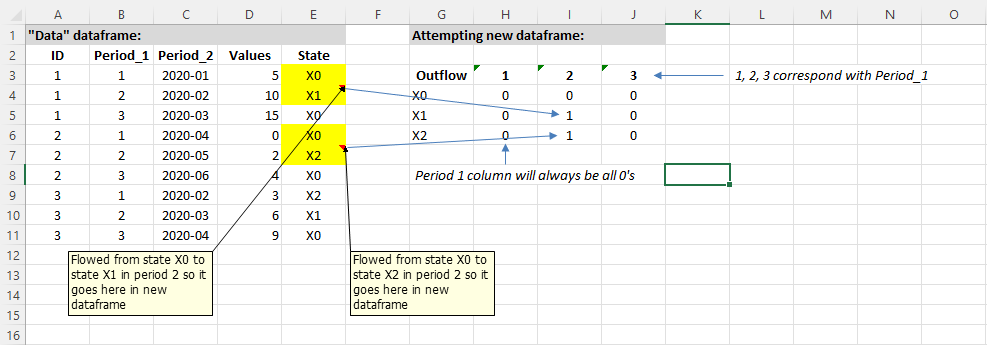
CodePudding user response:
dcast with drop = FALSE will create columns for every factor level of period_factor, so it you want columns 1 and 3 but not 4 in the result, we need to set the level of period_factor to include 1 and 3 but not 4 (and keep it factor class!). I've simplified the creation of period_factor too, lapply and .SDcols would only be needed if we were applying this to multiple columns:
dcast(
setDT(data)[, OutflowState := factor(shift(State, type = c("lead"))), by = ID]
[, period_factor := factor(Period_1 1, levels = seq(1, max(Period_1)))],
OutflowState ~ period_factor, length,
value.var = "Values",
subset = .(State == "X0" ),
drop = FALSE
)
# OutflowState 1 2 3
# 1: X0 0 0 0
# 2: X1 0 1 0
# 3: X2 0 1 0
CodePudding user response:
We can take the state_inflow() function from my answer to the question How to use data.table to build a new dataframe showing inflows into a specified transition state based on the value of an element in a prior row? and turn it into a state_outflow() function:
state_inflow <- function(mydat, target_state) {
dcast(
setDT(mydat)[, Previous_State := shift(State, fill = target_state), by = ID],
factor(Previous_State) ~ factor(Period_1), length, value.var = "Values",
subset = .(State == target_state), drop = FALSE
)
}
by interchanging the roles of State and Previous_State in dcast() :
state_outflow <- function(mydat, target_state) {
dcast(
setDT(mydat)[, Previous_State := shift(State), by = ID],
factor(State) ~ factor(Period_1), length, value.var = "Values",
subset = .(Previous_State == target_state), drop = FALSE
)
}
In addition, no fill value is supplied to the shift() function. So, the previous state of the first period of each ID is NA.
Calling the function gives the expected result:
state_outflow(data, "X0")
State 1 2 3 1: X0 0 0 0 2: X1 0 1 0 3: X2 0 1 0
There is no need to tweak factor levels.
