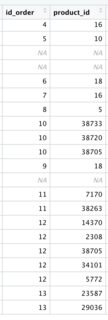
I'm preparing a master's degree project and stuck with basic data manipulation. I'm importing several data from the Prestashop database to R, one of those is a data frame with carts IDs and products included in it (see below).
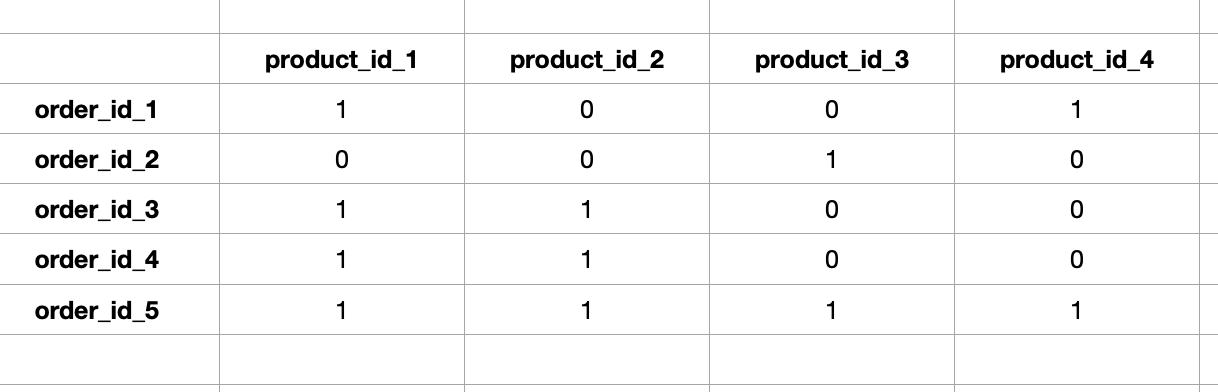
What I want to do is to create a matrix that will reflect the same data but in the easiest way as a matrix, here's a draft of the most desirable look:
Any hints on how the code should look? Thank you in advance for any help!
EDIT:
Code sample (dataframe):
x <- data.frame (order_id = c("12", "13","13","13","14","14","15","16"),
product_id = c("123","123","378","367","832","900",NA,"378"))
SOLUTION:
xtabs is good, but when it comes to NA values it skips the line in the results. There's an option to force addNA=TRUE, but it adds the NA 'column' and counts the NA as 1 (see below)
y <- xtabs(formula = ~., data = x)
Output - example 1 (addNA=FALSE):
product_id
order_id 123 367 378 832 900
12 1 0 0 0 0
13 1 1 1 0 0
14 0 0 0 1 1
16 0 0 1 0 0
Output - example 2 (addNA=TRUE):
product_id
order_id 123 367 378 832 900 <NA>
12 1 0 0 0 0 0
13 1 1 1 0 0 0
14 0 0 0 1 1 0
15 0 0 0 0 0 1
16 0 0 1 0 0 0
The igraph approach seems to be more accurate.
CodePudding user response:
You are looking for creating an adjacency matrix from a bipartite network from which you have the nodes list. You can directly use the package igraph to create the adjacency matrix from the node list and simplify it.
From x:
order_id product_id
1 12 123
2 13 123
3 13 378
4 13 367
5 14 832
6 14 900
7 15 <NA>
8 16 378
graph_from_dataframe <- igraph::graph.data.frame(x)
adjacency_matrix <- igraph::get.adjacency(graph_from_dataframe, sparse = FALSE)
# removing redundant entries
adjacency_matrix <- adj[rownames(adj) %in% x$order_id, colnames(adj) %in% x$product_id]
123 378 367 832 900
12 1 0 0 0 0
13 1 1 1 0 0
14 0 0 0 1 1
15 0 0 0 0 0
16 0 1 0 0 0
More resources on this SO question and this RPubs blog post.