The below code is being used to analysis a cvs file and at the moment im trying to remove the columns of the array which are not in my check_list. This only checks the first row and if the first row of the particular column doesnt belong to the check_list it removes the entire column. But this error keeps getting thrown and not sure how to avoid it.
def load_metrics(filename):
"""opens a cvs file and returns stuff"""
check_list = ["created_at","tweet_ID","valence_intensity","anger_intensity","fear_intensity","sadness_intensity","joy_intensity","sentiment_category","emotion_category"]
file=open(filename)
data = []
for lin in file:
lin = lin.strip()
lin = lin.split(",")
data.append(lin)
for col in range(len(data[0])):
if np.any(data[0][col] not in check_list) == True:
data[0]= np.delete(np.array(data), col, 1)
print(col)
return np.array(data)
The below test is being used on the code too, if that helps
Test
data = load_metrics("covid_sentiment_metrics.csv")
print(data[0])
Results
['created_at' 'tweet_ID' 'valence_intensity' 'anger_intensity'
'fear_intensity' 'sadness_intensity' 'joy_intensity' 'sentiment_category'
'emotion_category']
CodePudding user response:
Change your load_metrics function to:
def load_metrics(filename):
check_list = ["created_at","tweet_ID","valence_intensity","anger_intensity",
"fear_intensity","sadness_intensity","joy_intensity","sentiment_category",
"emotion_category"]
data = []
with open(filename, 'r') as file:
for lin in file:
lin = lin.strip()
lin = lin.split(",")
data.append(lin)
arr = np.array(data)
colFilter = []
for col in arr[0]:
colFilter.append(col in check_list)
return arr[:, colFilter]
I introduced the following corrections:
- Use with to automatically close the input file (your code fails to close it).
- Create a "full" Numpy array (all columns) after the data has been read.
- Compute colFilter list - which columns are in check_list.
- Return only filtered columns.
CodePudding user response:
This code does not include checks related to reading a file or a broken data structure, so that the main idea is more or less clear. So, here I assume that a csv-file exists and has at least 2 lines:
import numpy as np
def load_metrics(filename, check_list):
"""open a csv file and return data as numpy.array
with columns from a check list"""
data = []
with open(filename) as file:
headers = file.readline().rstrip("\n").split(",")
for line in file:
data.append(line.rstrip("\n").split(","))
col_to_remove = []
for col in reversed(range(len(headers))):
if headers[col] not in check_list:
col_to_remove.append(col)
headers.pop(col)
data = np.delete(np.array(data), col_to_remove, 1)
return data, headers
Quick testing:
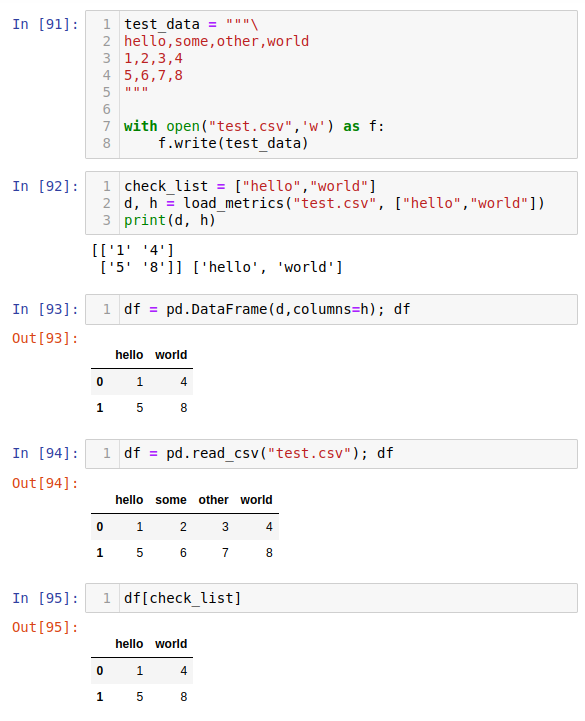
test_data = """\
hello,some,other,world
1,2,3,4
5,6,7,8
"""
with open("test.csv",'w') as f:
f.write(test_data)
check_list = ["hello","world"]
d, h = load_metrics("test.csv", ["hello","world"])
print(d, h)
Expected output:
[['1' '4']
['5' '8']] ['hello', 'world']
Some details:
- Instead of
np.any(data[0][col] not in check_list) == Truewould be enoughdata[0][col] not in check_list - Stripping with default parameters is not good as far as you can delete meaningful spaces.
- Do not delete anything while looping forward. But we can do it (with some reservations) while looping backward.
check_listis better as a parameter.- Separate data and headers as they may have different types.
- In your case it is better to use
pandas.read_csv, see the picture below.