I cannot figure out how I should tackle this problem:
I have dataframe of measurements from cancer-treatments that i want to 'merge' pairwise using coalesce(). But the frame contains 100 columns so i want to use some kind of function or loop.
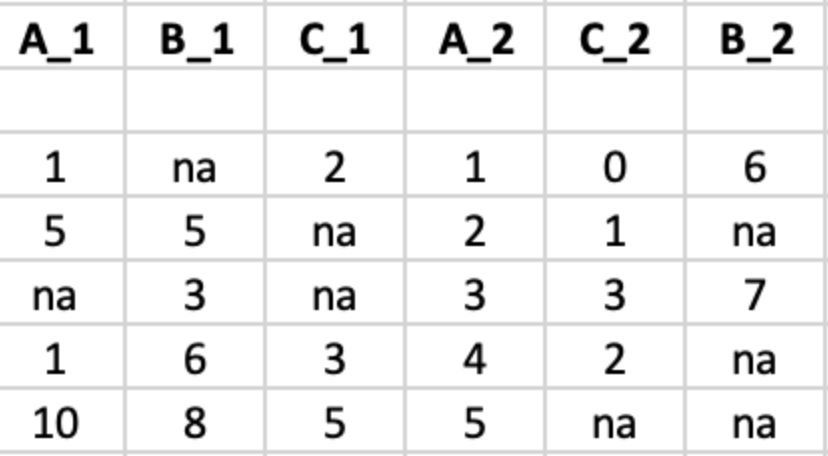
Here's what my dataframe looks like
I have a vector with the names of the treatments: drugs <- c(A, C, B, D etc)
the columns represent multiple measurements from a drug treatment. I want to merge the columns pairwise: A_1 A_2 into A (new column), B_1 etc
This code works: df <- df %>% mutate(A = coalesce(A_1, A_2).
But the frame has 100 columns so i want to use some kind of function or loop, using the value from the vector with drugnames. From each drug there are 2 columns but they are not in the correct order, so i cannot use numbering, i have to use the name of the column. But when i put that into a function it doesn't work
One addition: i would like to have the resulting columns (A,B,C) etc added to the frame.
How should i proceed?
thanks!!
CodePudding user response:
Try this approach:
library(tidyverse)
tribble(
~A1, ~B1, ~C1, ~A2, ~C2, ~B2,
1, NA, 2, 1, 0, 6,
5, 5, NA, 2, 1, NA,
NA, 3, NA, 3, 3, 7,
1, 6, 3, 4, 2, NA,
10, 8, 5, 5, NA, NA
) |>
mutate(row = row_number()) |>
pivot_longer(-row, names_to = c("prefix", "suffix"),
names_pattern = "(\\w)(\\d)") |>
pivot_wider(names_from = suffix, names_prefix = "part_",
values_from = value) |>
mutate(coalesced = coalesce(part_1, part_2)) |>
select(- starts_with("part_")) |>
pivot_wider(names_from = prefix, values_from = coalesced) |>
select(-row)
#> # A tibble: 5 × 3
#> A B C
#> <dbl> <dbl> <dbl>
#> 1 1 6 2
#> 2 5 5 1
#> 3 3 3 3
#> 4 1 6 3
#> 5 10 8 5
Created on 2022-06-04 by the reprex package (v2.0.1)
CodePudding user response:
Here is an option with split.default - split the data into chunks of data based on the column names pattern, then use coalesce by looping over the list
library(dplyr)
library(stringr)
library(purrr)
df1 %>%
split.default(str_remove(names(.), "\\d $")) %>%
map_dfc(~ exec(coalesce, !!!.x))
-output
# A tibble: 5 × 3
A B C
<dbl> <dbl> <dbl>
1 1 6 2
2 5 5 1
3 3 3 3
4 1 6 3
5 10 8 5
data
df1 <- structure(list(A1 = c(1, 5, NA, 1, 10), B1 = c(NA, 5, 3, 6, 8
), C1 = c(2, NA, NA, 3, 5), A2 = c(1, 2, 3, 4, 5), C2 = c(0,
1, 3, 2, NA), B2 = c(6, NA, 7, NA, NA)), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -5L))