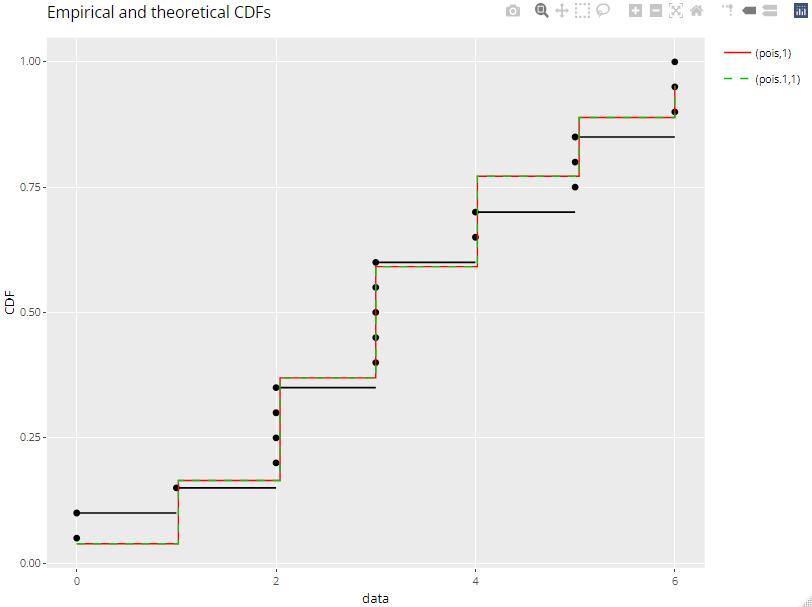
I do have a similar behavior than 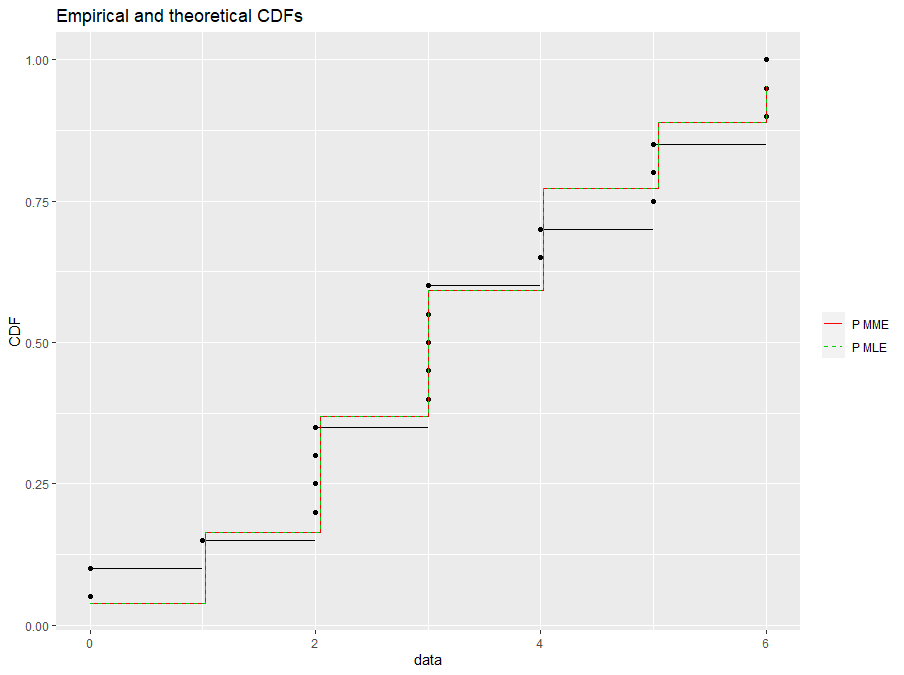
Code :
library(fitdistrplus)
set.seed(123)
X <- rpois(20,3)
fit.p.mme <- fitdist(data = X, distr = "pois", method = "mme")
fit.p.mle <- fitdist(data = X, distr = "pois", method = "mle")
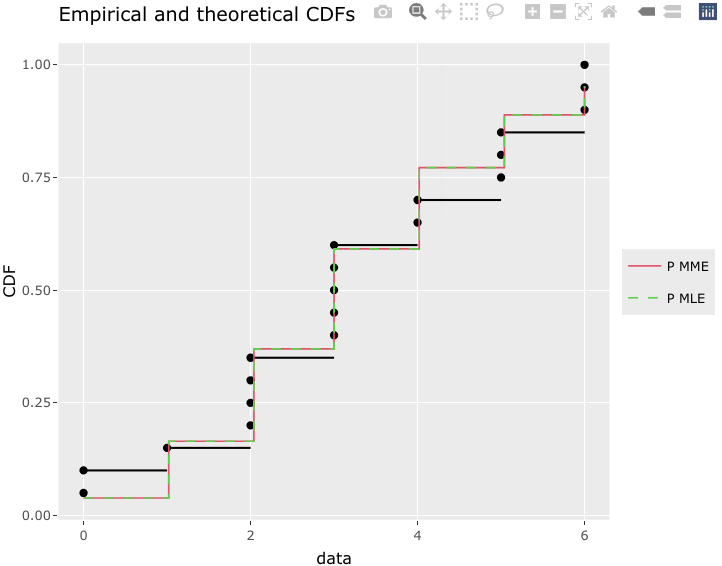
g1 <- cdfcomp(list(fit.p.mme, fit.p.mle), addlegend = TRUE, legendtext = c("P MME", "P MLE"), plotstyle = "ggplot")
g1 <- g1 theme_gray()
g1
g1 %>% ggplotly()
Tryed to edit labels in g1$data$ind without success.
CodePudding user response:
You can change the names manually in a Plotly build.
To find out what names came from where I built the plot and ran lapply to output the names and legend groups. In plotly, discrete color assignments will get their own trace. So the name of the trace is typically the name in the legend when it's built.
I didn't need to call ggplotly() first, either.
g2 <- plotly_build(g1)
invisible(
lapply(1:length(g2$x$data),
function(j) {
message(paste0(j, " ", g2$x$data[[j]]$name, " & ",
g2$x$data[[j]]$legendgroup))
})
)
# 1 &
# 2 &
# 3 &
# 4 &
# 5 (pois,1) & (pois,1)
# 6 (pois.1,1) & (pois.1,1)
So there are 6 traces, and these are their names (less the ampersand that I pasted in there).
You can change these names directly.
g2$x$data[[5]]$name <- "P MME"
g2$x$data[[5]]$legendgroup <- "P MME"
g2$x$data[[6]]$name <- "P MLE"
g2$x$data[[6]]$legendgroup <- "P MLE"
If you wanted the legend centered on the y-axis with the plot background color, you can do that here, too.
g2$x$layout$legend$y <- .5 # if you want to move the legend to the middle
g2$x$layout$legend$bgcolor <- g2$x$layout$plot_bgcolor # to use the same bg color
g2 # let's see it
It's definitely not the same, but it's closer.