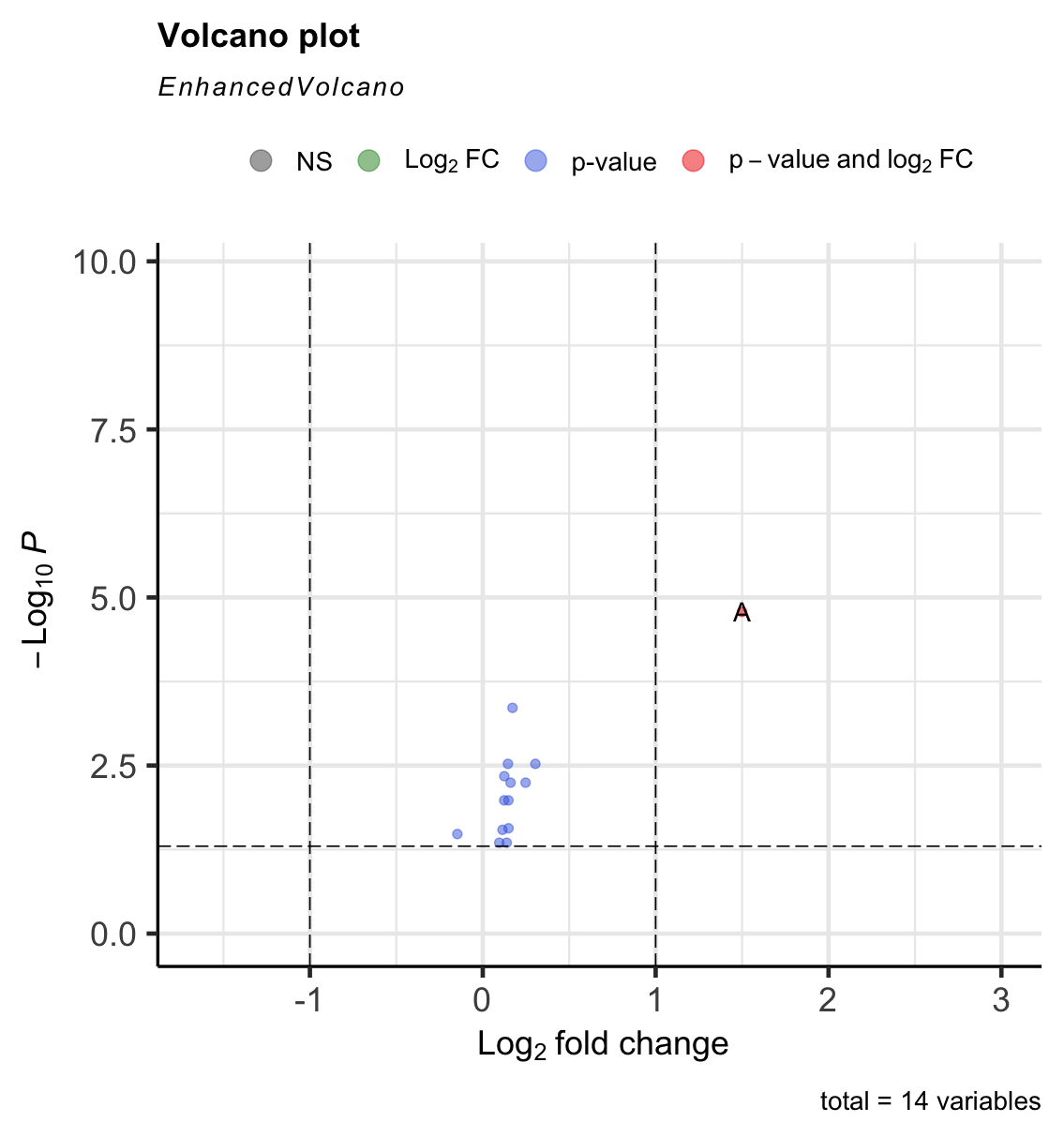
I am using the following function in R to develop a simple volcano plot:
EnhancedVolcano(all_genes, x = "logFC", y = "adjust.p.value", lab = all_genes$Gene.ID,
pCutoff = 10e-2, FCcutoff = 1)
I would like my pCutoff line to appear to represent p = 0.05 which on a log scale for this figure would appear as 1.3 on the y-axis. However, changing "10e-2" to say "10e-2.5" generates an error
Error: unexpected numeric constant in: "EnhancedVolcano(all_genes, x = "logFC", y = "adjust.p.value", lab = all_genes$Gene.ID, pCutoff = 10e-2.5"
Any suggestions on how I can get a horizontal p-value cut-off line at exactly 1.3 (currently appears at 1.2). Here is some reproducible data:
structure(list(X = 1:14, Gene.ID = c("A", "B", "C", "D", "E", "F", "G", "H", "I", "J", "K", "L", "M", "N"), logFC = c(1.5, 0.17212922, 0.145542174, 0.304348578, 0.124636936, 0.247841044, 0.160818268, 0.123741518, 0.148530876, 0.148960225, 0.114135472, -0.147118359, 0.095549291, 0.138521594), AveExpr = c(5.426424957, 4.289728233, 4.901134193, 4.742864705, 5.447030699, 4.539641767, 4.650750102, 5.901020922, 5.365944907, 5.818788787, 4.837214384, 7.017656548, 4.531897822, 5.192294452), t = c(6.15098624, 5.452898247, 4.979246654, 4.949519834, 4.818043279, 4.73403717, 4.701937811, 4.522692175, 4.518518374, 4.281900066, 4.247981727, -4.194421592, 4.10350597, 4.088357671), p.value = c(1.27e-09, 6.8e-08, 7.99e-07, 9.26e-07, 1.77e-06, 2.65e-06, 3.09e-06, 7.13e-06, 7.27e-06, 2.1e-05, 2.44e-05, 3.07e-05, 4.53e-05, 4.83e-05), adjust.p.value = c(1.64e-05, 0.000438854, 0.002987004, 0.002987004, 0.004558267, 0.005687325, 0.005687325, 0.010422933, 0.010422933, 0.027128901, 0.028601707, 0.033061438, 0.04452146, 0.04452146), B = c(11.2786109, 7.664706936, 5.439886439, 5.306497286, 4.725465519, 4.361868581, 4.224515919, 3.473656504, 3.45649938, 2.508304771, 2.376338878, 2.169980059, 1.825392322, 1.76867543)), class = "data.frame", row.names = c(NA, -14L))
CodePudding user response:
I think you want the following code where the p-value is calculated like p=10^-s where s is your 1.3 like this:
library(EnhancedVolcano)
EnhancedVolcano(all_genes, x = "logFC", y = "adjust.p.value", lab = all_genes$Gene.ID,
pCutoff = 10^-1.3, FCcutoff = 1)
Output: