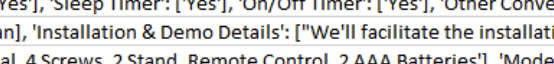
I have a blurb of text in 500 rows in 1 of the many columns as below:
{'Smart Remote': ['Yes'], 'Color Screen': ['Yes'], 'RF Capable': ['Yes'], 'Internet Access': ['Yes']}
I need to append number of columns like this:
Smart Remote : Yes, Color Screen : Yes, RF Capable : Yes, Internet Access : Yes
Note that entries in {} are random in order, with few unique entries per row
Edit : Added image snip of data set

CodePudding user response:
Sample data, I hope this is close enough to what you have:
dat <- structure(list(id = 1:2, txt = c("{'Smart Remote': ['Yes'], 'Color Screen': ['Yes'], 'RF Capable': ['Yes'], 'Internet Access': ['Yes']}", "{'RF Capable': ['No'], 'Internet Access': ['No'],'Smart Remote': ['No'], 'Color Screen': ['No']}")), class = "data.frame", row.names = c(NA, -2L))
(Note that I varied the data and the order of the fields.)
dplyr
library(dplyr)
# library(tidyr) # unnest
# library(jsonlite) # fromJSON
dat %>%
mutate(data = lapply(txt, \(x) data.frame(jsonlite::fromJSON(gsub("'", '"', x)), check.names = FALSE))) %>%
tidyr::unnest(data)
# # A tibble: 2 x 6
# id txt `Smart Remote` `Color Screen` `RF Capable` `Internet Access`
# <int> <chr> <chr> <chr> <chr> <chr>
# 1 1 {'Smart Remote': ['Yes'], 'Color Screen': ['Yes'], 'RF Capable': ['Yes'], 'Internet Access': ['Yes']} Yes Yes Yes Yes
# 2 2 {'RF Capable': ['No'], 'Internet Access': ['No'],'Smart Remote': ['No'], 'Color Screen': ['No']} No No No No
data.table
library(data.table)
# library(jsonlite) # fromJSON
DT <- as.data.table(dat)
DT[, c(.SD, rbindlist(lapply(txt, \(x) jsonlite::fromJSON(gsub("'", '"', x))), use.names = TRUE, fill = TRUE))]
# id txt Smart Remote Color Screen RF Capable Internet Access
# <int> <char> <char> <char> <char> <char>
# 1: 1 {'Smart Remote': ['Yes'], 'Color Scr... Yes Yes Yes Yes
# 2: 2 {'RF Capable': ['No'], 'Internet Acc... No No No No
CodePudding user response:
If the data is supposed to be JSON, there is an error in it: quotes around a JSON object should always be double quotes, while this data uses mostly single quotes. As such, R's JSON libraries can't seem to load it.
We can't just replace all single quotes with double quotes: this data uses double quotes whenever there are single quotes inside a string; we need to make sure that those single quotes remain untouched:
Replacing ' with "
From the excerpt we can distill the task of this transformation:
in: {'Details':["We'll facilitate"]}
out: {'Details':['We\'ll facilitate']}
We don't know yet what the input looks like when two types of quotes are present in a string:
{'Details':["We'll facilitate \"everything\" "]}or{'Details':['We\'ll facilitate "everything" ']}- or maybe some completely different way of escaping.
For now I'll handle just the first transformation, and ignore the second.
The function below walks through a string, remembers if we're inside single or double quotes, and changes single quotes to double quotes accordingly:
repairString <- function(string) {
cat('in:', string, '\n')
inside <- FALSE # store if we're inside single / double quotes or none at all
i <- 1 # string index
while(i <= nchar(string)) {
chr <- substr(string, i, i) # get single character
if(chr %in% c("'", '"')) { # check if ' or "
if(isFALSE(inside)) { # not yet inside quotes: we're entering them
inside <- chr
swap <- TRUE
} else if(inside == chr) { # inside the same quotes: we're exiting them now
inside <- FALSE
swap <- TRUE
} else { # inside the other type of quote: we need to skip this one
swap <- FALSE
}
# swap ' for " if required
if(swap & chr == "'") string <- paste0(substr(string, 1, i-1), '"', substr(string, i 1, nchar(string)))
}
i <- i 1
}
cat('out:',string,'\n\n')
return(string)
}
Example:
> repairString("{'Details':[\"We'll facilitate\"]}")
in: {'Details':["We'll facilitate"]}
out: {"Details":["We'll facilitate"]}
For this small example dataset I mocked up, we can use this function to repair all strings and store them in the data frame:
df <- structure(list(features = c("{'Smart Remote': ['Yes'], 'Color Screen': ['Yes'], 'RF Capable': ['Yes'], 'Internet Access': ['Yes']}",
"{'Hypergalactic': ['No'], 'Waterproof': ['No','Well, sometimes'], 'Good With Dogs': [\"No they're very bad with dogs\"]}"
)), class = "data.frame", row.names = c(NA, -2L))
df$features.fixed <- lapply(df$features, repairString)
Printing the data
Now that the strings are repaired, they can be read, parsed, stored and printed:
# iterate over all rows
lapply(df$features.fixed, function(row) {
# parse JSON
row_data <- jsonlite::fromJSON(row, simplifyVector = F)
# join multiple values with ' / '
row_data <- mapply(function(x) paste0(unlist(x), collapse=' / '), row_data)
# paste names and values together
text <- sapply(seq_along(row_data), function(x) paste0(names(row_data)[x], ': ', row_data[x]) )
# paste multiple 'name: value' strings together
paste0(text, collapse = '; ')
}) -> df$features.string
> df$features.string
[[1]]
[1] "Smart Remote: Yes; Color Screen: Yes; RF Capable: Yes; Internet Access: Yes"
[[2]]
[1] "Hypergalactic: No; Waterproof: No / Well, sometimes; Good With Dogs: No they're very bad with dogs"