I have an enormous "omics" dataset containing three different experiments: df$method == Mut, Spy, VAR
method a b c d
1 Mut 12.3 NA NA 17.5
2 Spy 13.5 NA NA NA
3 VAR 13.2 19.6 11.1 NA
4 Mut NA NA NA NA
5 Spy NA NA NA 19.9
6 VAR NA 20.1 18.6 NA
Using dplyr, how can I reduce the matrix so it only contains rows where df$method == VAR has values (at least one value)? I.e., where all values in a, b, c, d ... is NA for df$method == Mut, Spy.
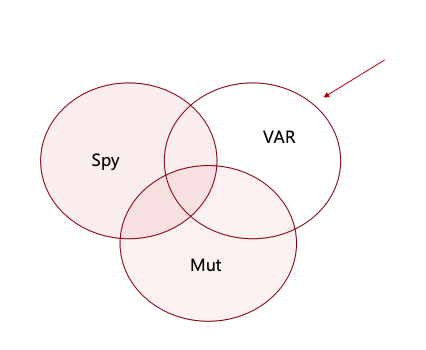
Shown on a Venn Diagramm, values that fits in the white area, are of interest.
So, the expected output from df would be:
> df
method b c
1 VAR 19.6 11.1
2 VAR 20.1 18.6
Data
df <- structure(list(method = c("Mut", "Spy", "VAR", "Mut", "Spy",
"VAR"), a = c(12.3, 13.5, 13.2, NA, NA, NA), b = c(NA, NA, 19.6,
NA, NA, 20.1), c = c(NA, NA, 11.1, NA, NA, 18.6), d = c(17.5,
NA, NA, NA, 19.9, NA)), class = "data.frame", row.names = c(NA,
-6L))
CodePudding user response:
dplyr option to first filter the method and then select the columns with no NA's like this:
df <- structure(list(method = c("Mut", "Spy", "VAR", "Mut", "Spy",
"VAR"), a = c(12.3, 13.5, 13.2, NA, NA, NA), b = c(NA, NA, 19.6,
NA, NA, 20.1), c = c(NA, NA, 11.1, NA, NA, 18.6), d = c(17.5,
NA, NA, NA, 19.9, NA)), class = "data.frame", row.names = c(NA,
-6L))
library(dplyr)
library(dplyr)
df %>%
filter(method == "VAR") %>%
select_if(~!any(is.na(.)))
#> method b c
#> 1 VAR 19.6 11.1
#> 2 VAR 20.1 18.6
Created on 2022-07-06 by the reprex package (v2.0.1)
CodePudding user response:
Here is a base R way. Use logical indices to get the rows where method == "VAR" and the columns where the other rows, the rows with method is equal to "Spy" or "Mut" are all NA.
df <- structure(list(
method = c("Mut", "Spy", "VAR", "Mut", "Spy","VAR"),
a = c(12.3, 13.5, 13.2, NA, NA, NA),
b = c(NA, NA, 19.6,NA, NA, 20.1),
c = c(NA, NA, 11.1, NA, NA, 18.6),
d = c(17.5,NA, NA, NA, 19.9, NA)),
class = "data.frame", row.names = c(NA,-6L))
i_row <- df$method == "VAR"
i_col <- colSums(is.na(df[!i_row, -1])) == nrow(df[!i_row,])
df[i_row, c(TRUE, i_col)]
#> method b c
#> 3 VAR 19.6 11.1
#> 6 VAR 20.1 18.6
Created on 2022-07-06 by the reprex package (v2.0.1)