I would like to present some 3D spatial data with Python, but with the geometry of the setup plotted for each direction. So for the XY plane, I would have at z=0 the view from above, same for XZ and YZ.
I saw that other posts asked a similar question, but here, the data would not be coordinates and colors, but rather a real picture.
My idea, based on other posts was the following:
import matplotlib.pyplot as plt
from mpl_toolkits import mplot3d
import numpy as np
import matplotlib.image as mpimg
fig = plt.figure()
fig.add_subplot(111, projection="3d")
# 3D data
X = np.random.uniform(0,100,100)
Y = np.random.uniform(0,100,100)
Z = np.random.uniform(0,100,100)
values = np.random(100)
# XY
img = mpimg.imread('/home/geometryXY')
imgplot = plt.imshow(img, zdir='x', aspect='auto')
# XZ
img = mpimg.imread('/home/geometryXZ')
imgplot = plt.imshow(img, zdir='y', aspect='auto')
# YZ
img = mpimg.imread('/home/geometryYZ')
imgplot = plt.imshow(img, zdir='z', aspect='auto')
plt.show()
Any image file would work. Indeed, for now, what interests me is to see if we can put a 2D image on a 3D axis and overlap a 3D plot on it.
At least the idea would be that. However, the 'zdir' option does not seem to exist. Would you have any tip?
Thanks
EDIT:
Based on blunova's answer, I changed several things. First I added where I want to see the plot with the val argument, and scaling parameters for each axis xlim and ylim. To make it work, for some reason, I also had to swap the x and y axes of the imread object and to flip the y axis.
Several problems are still here.
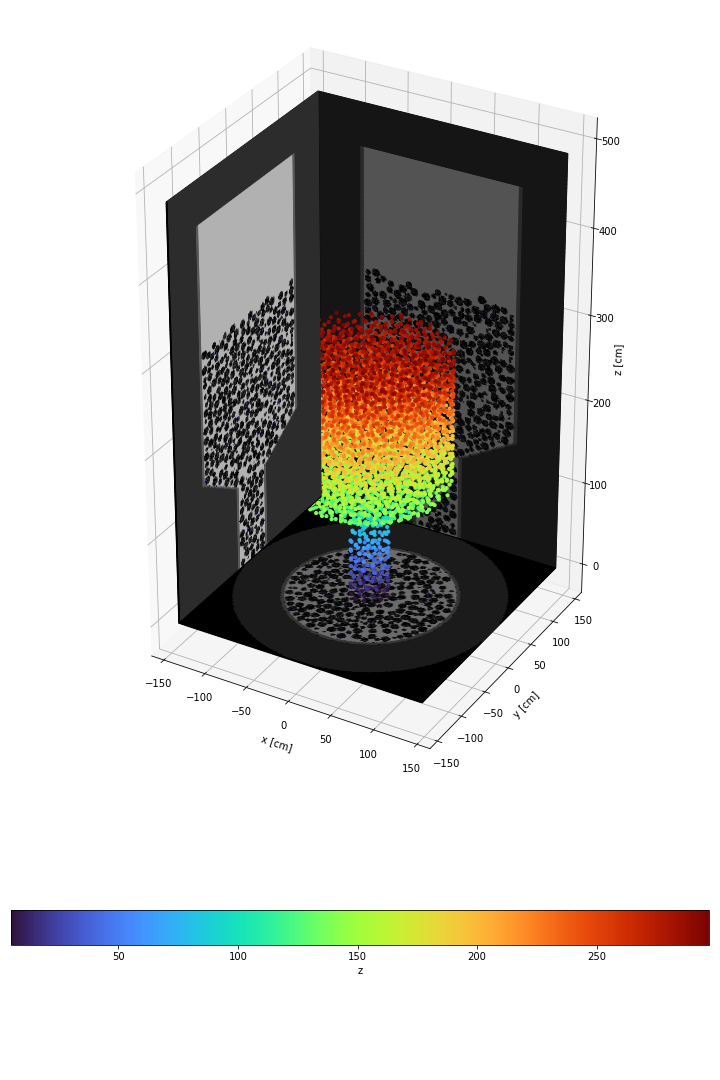
The first one is that the process is very slow. Showing the 2D picture with imshow would take less few seconds (images are 3000*5000 pixels large), but here I stopped the function after few minutes because it was too slow. I therefore changed the rstride and cstride, and mutliplied them by 15, which is huge (does it mean I reduce my quality by 15^2?) just to make it barely ok (2 minutes to plot 3 images). What I don't understand why there is such a difference between the normal imshow and the plot_surface routines.
The second is that the images "cut" the 3D data in foreground, even though I added the 3D plot afterwards, and even if the zorder for the images is set to -1000. The 3D data should show a full cylinder, and no value is lower than the -144 value shown in the code.
There also seems to be a problem with the lighting which is strange.
For your reference, here is the current state of this plot.
def plot_image(ax, image, axis, xlim, ylim, val, rstride=15, cstride=15):
array = plt.imread(image)
array = np.swapaxes(array, 0, 1)
array = np.flip(array, 1)
step_x, step_y = np.diff(xlim) / array.shape[0], np.diff(ylim) / array.shape[1]
x_1 = np.arange(xlim[0], xlim[1], step_x)
y_1 = np.arange(ylim[0], ylim[1], step_y)
y_1, x_1 = np.meshgrid(y_1, x_1)
vals = np.ones((array.shape[0], array.shape[1]))*val
if axis == "x":
ax.plot_surface(vals, x_1, y_1, rstride=rstride, cstride=cstride, facecolors=array, zorder=-1000)
elif axis == "y":
ax.plot_surface(x_1, vals, y_1, rstride=rstride, cstride=cstride, facecolors=array, zorder=-1000)
elif axis == "z":
ax.plot_surface(x_1, y_1, vals, rstride=rstride, cstride=cstride, facecolors=array,zorder=-1000)
fig = plt.figure(figsize=(10,15))
fig.add_subplot(111, projection="3d")
# Plot geometry in background
plot_image(plt.gca(), '/home/yryves/serpent_cases/domain_decomposition_dvlpt/test_stl2/Plots/input_geom1_bu1.png','x', xlim=[-144,144], ylim=[0,487.528], val=-144)
plot_image(plt.gca(), '/home/yryves/serpent_cases/domain_decomposition_dvlpt/test_stl2/Plots/input_geom2_bu1.png','y', xlim=[-144,144], ylim=[0,487.528], val=144)
plot_image(plt.gca(), '/home/yryves/serpent_cases/domain_decomposition_dvlpt/test_stl2/Plots/input_geom3_bu1.png','z', xlim=[-144,144], ylim=[-144,144], val=0)
# 3D data plot
pbed.plot3D('z', new_fig=False)
plt.savefig('geometry.png')
CodePudding user response: