I'm trying to plot data from several columns of a dataframe. Each column represents the y value and for each of those, the x-value range from 0 to 100. When trying to graph this data, they y-axis looks like a giant mess(there are 12 columns). If I can set a minimum y value of 0 and a max of 1000 or so, things should workout. How do I do this? Also how do I extend the x axis so that I may add a legend next to it(How do I do that here?) ggplot is not being recognized by my Rstudio so I used this code
plot(x-axis, y1)
par(new=T)
plot(x-axis, y2, col="darkcyan")
par(new=T)
plot(x-axis, y3, col="green")
par(new=T)
plot(x-axis, y4, col="orange")
par(new=T)
...
plot(x-axis, y12, col="blue")
par(new=T)
I also wrote this other code to do the same thing.
matplot(x-axis,
cbind(aaaa,aaab,aaac,aaad,aaae,aaaf,aaag,aaah,
aaai,aaaj, aaak,aaal),type="l",col=c("red", "blue",
"green", "cyan", "darkcyan", "azure",
"darkkhaki","aquamarine", "brown1","deeppink",
"deepskyblue1", "blueviolet"), xlab="Return Period
(Years)", ylab="Return Years (mm)" )
legend("topright", colnames(dfreturnplot2), col=12,
cex=0.8, fill=seq_len(12))
Here, the y-axis looks fine but legend is interfering with the visibility of the graph. I can't just make a longer x-dimension by lengthening x-axis so what I should do?
CodePudding user response:
Extend the ylim by a factor say *1.1 and make a frameless horizontal legend.
matplot(m, type='l', lty=1, ylim=c(min(m), max(m)*1.1))
legend("topleft", colnames(m), col=12, cex=0.8, fill=seq_len(12), horiz=TRUE, bty='n')
If you have even more items, there's also a ncol= argument in legend to present them in a multicolumn matrix, see ?legend.
matplot(m, type='l', lty=1, ylim=c(min(m), max(m)*1.1))
legend("topleft", LETTERS[1:20], col=12, cex=0.8, fill=seq_len(20), ncol=10, bty='n')
You may also extend the xlim and leave the frame on, this is really very flexible.
matplot(m, type='l', lty=1, xlim=c(0, nrow(m)*1.25))
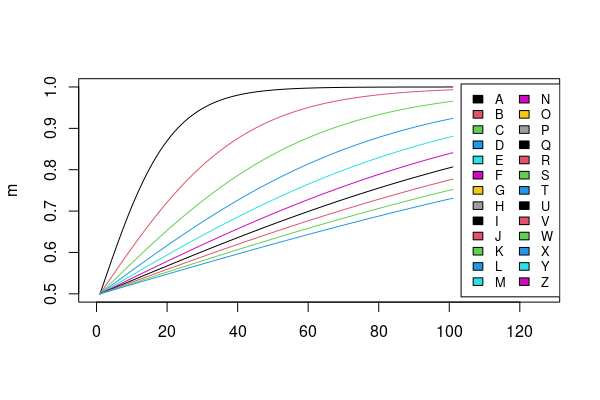
legend("right", LETTERS[1:26], col=12, cex=0.8, fill=seq_len(20), ncol=2)
Data:
m <- sapply(1:10, \(x) plogis(seq(0, 10, .1)/x)) |> `colnames<-`(LETTERS[1:10])
CodePudding user response:
I urge trying to use ggplot2 here: it does legends very well and automatically shifts things around to make everything fit.
ggplot is not being recognized by my Rstudio so I used this code
Try install.packages("ggplot2"); if it fails, some quick research should inform how to get it installed.
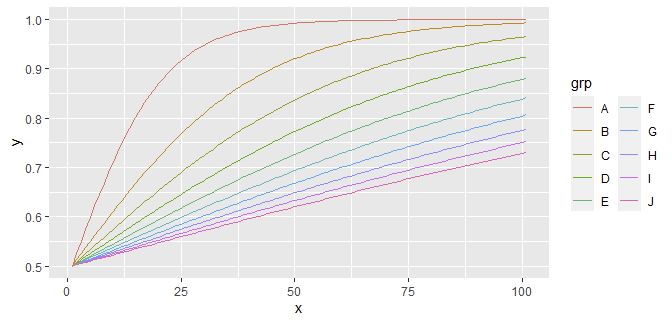
Using @jay.sf's sample data, we just need to melt it and ggplot2 does the rest:
library(ggplot2)
transform(m, x = seq_len(nrow(m))) |>
reshape2::melt("x", variable.name = "grp", value.name = "y") |>
ggplot(aes(x, y, color = grp))
geom_line()
guides(color = guide_legend(ncol = 2))
ggplot2 allows you to customize a lot, including multi-column legends (as shown, defaults to 1), removing legend titles, cleaning up the background/theme (add theme_bw()), varying color/size/linetype/... by group/line/..., etc. Learn its semantics once and you will forever benefit from its ability to control great-looking plots (versus base graphics where you have to wrestle control for large legends manually).