I would like some help in creating a dataframe in R for per predicted chances.
I have a dataset of 156 patients, 39 without disease and 117 with disease and all patients have a predicted chance of disease (0,00-1). To determine a cut-off point I would like to create a dataset in which per increase of 1% chance the amount of with and without disease is shown.
So a dataset with 101 obs and 3 variables (percent chance, amount of patients with disease, amount of patients without disease)
I created the following loop, but it results in 287 observations.
Valid2$disease_T <- Valid2$disease_present
Valid2$disease_F <- ifelse(Valid2$disease_present == F, T, F)
plot2 <- data.frame()
for (i in 0:100) {
per <- i
amount <- Valid2 %>%
select(disease_T, disease_F, predicted) %>%
filter (predicted >= (i/100)) %>%
count(disease_T, disease_F)
plot2 <- rbind(plot2, per, amount)
}
dput of Valid2
structure(list(disease_T = c(TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, TRUE, TRUE, FALSE,
FALSE, FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, TRUE, FALSE, FALSE,
TRUE, FALSE, FALSE, FALSE, TRUE), disease_F = c(FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE,
TRUE, TRUE, FALSE, FALSE, TRUE, TRUE, TRUE, TRUE, FALSE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, FALSE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE,
TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, FALSE, TRUE, TRUE,
FALSE, TRUE, TRUE, TRUE, FALSE), predicted = c(0.839047189743821,
0.967837763160834, 0.786317285878715, 0.675276246104666, 0.989345368556793,
0.555983868574255, 0.990942898545037, 0.811649758224556, 0.98426150620371,
0.736827268936253, 0.979475139855437, 0.774123207779965, 0.957903428160418,
0.91999882380089, 0.942305155149908, 0.991921678870239, 0.991594114313076,
0.849292946424851, 0.97569066014882, 0.884909080808976, 0.979013574591358,
0.998862491367365, 0.923394431637592, 0.885001406361028, 0.877973037376933,
0.728550121991711, 0.999833278501221, 0.897790177969505, 0.979220428963593,
0.999499362617635, 0.954494527800288, 0.961765702221913, 0.769280528680882,
0.882613166009711, 0.986217524084092, 0.954192408993744, 0.872914657137128,
0.531299550904597, 0.893933172484704, 0.480533848683641, 0.993094398777802,
0.824802835501908, 0.947570508969646, 0.980540528086075, 0.583395480728772,
0.822397774199589, 0.883912440796693, 0.594776303699779, 0.820790192098386,
0.798073877022062, 0.985966496663131, 0.897069576653414, 0.831132493632043,
0.852151002809375, 0.998435303514582, 0.953651187490499, 0.986591941625218,
0.955233427577533, 0.683855028732251, 0.996623760378059, 0.899794970651676,
0.89347885202678, 0.769929537527992, 0.795764657045519, 0.982146943465671,
0.776440313355684, 0.914849623160958, 0.970905780657116, 0.500682448346073,
0.863082837731646, 0.898528543977351, 0.961071059679792, 0.914764045084384,
0.740320486660299, 0.558067744300918, 0.924289384162258, 0.640027164915262,
0.991129364781095, 0.94097585658508, 0.948216610615068, 0.789283230550332,
0.965724911188744, 0.992147113271609, 0.990048301774303, 0.929031670004039,
0.909219568552839, 0.417005262010727, 0.954046684763806, 0.954662032660194,
0.592707632714186, 0.71736673909297, 0.815939418957414, 0.530573198572189,
0.411013385804287, 0.456143973275274, 0.98418041813448, 0.999743784673911,
0.748231061596753, 0.957616694642036, 0.936005342173473, 0.990966443212461,
0.998088129637225, 0.920524349831836, 0.995196908913598, 0.974348828931041,
0.973717852722492, 0.938994862330677, 0.533156741117527, 0.990726457099523,
0.513768273986449, 0.638444161218626, 0.858677012819686, 0.791287902868353,
0.588849209133098, 0.44811826975699, 0.508084588253886, 0.530249454616573,
0.488225901918474, 0.500562684131604, 0.317898539696961, 0.242177319047234,
0.609587716312933, 0.539440893692799, 0.355494594307387, 0.266099968050094,
0.723932395532802, 0.723938401792491, 0.53390474557177, 0.634097434175427,
0.775172549607967, 0.570928462844033, 0.522356812838135, 0.724635429147149,
0.610630883290112, 0.565371382980066, 0.285283409047343, 0.343302659495403,
0.816510539572742, 0.656765409452827, 0.626301633190735, 0.383723283273525,
0.594260652384327, 0.556639518107367, 0.418173506333977, 0.278806555045948,
0.516264516564629, 0.292843578210485, 0.576288502786766, 0.408152351764115,
0.650882387290395, 0.396480245419753, 0.834276346007703, 0.413110039326727,
0.561240114285867, 0.387299107426737, 0.620969313796766)), row.names = c(NA,
-156L), class = c("tbl_df", "tbl", "data.frame"))
CodePudding user response:
Note that you don't need to have both disease_T and disease_F
> (valid2$disease_T==!valid2$disease_F) %>% all
[1] TRUE
The below I think achieves what you're after.
percentages <- c(0:100)
valid3 <- valid2 %>% select(-disease_F) %>% # remove redundant column
mutate(roundedpercentage = ceiling(predicted*100)) %>%
rename(disease = disease_T) #rename to just disease
nb_sick <- sapply(percentages, function(x) sum(valid3$disease[valid3$roundedpercentage<=x]))
nb_healhy <- sapply(percentages, function(x) sum(!valid3$disease[valid3$roundedpercentage<=x]))
result <- data.frame(percentages, nb_sick, nb_healhy)
> result
percentages nb_sick nb_healhy
1 0 0 0
2 1 0 0
3 2 0 0
4 3 0 0
5 4 0 0
6 5 0 0
7 6 0 0
8 7 0 0
9 8 0 0
10 9 0 0
11 10 0 0
12 11 0 0
13 12 0 0
14 13 0 0
15 14 0 0
16 15 0 0
17 16 0 0
18 17 0 0
19 18 0 0
20 19 0 0
21 20 0 0
22 21 0 0
23 22 0 0
24 23 0 0
25 24 0 0
26 25 0 1
27 26 0 1
28 27 0 2
29 28 0 3
30 29 0 4
31 30 0 5
32 31 0 5
33 32 0 6
34 33 0 6
35 34 0 6
36 35 0 7
37 36 0 8
38 37 0 8
39 38 0 8
40 39 0 10
41 40 0 11
42 41 1 11
43 42 3 13
44 43 3 13
45 44 3 13
46 45 3 14
47 46 4 14
48 47 4 14
49 48 4 14
50 49 5 15
51 50 5 15
52 51 7 16
53 52 8 17
54 53 8 18
55 54 11 21
56 55 11 21
57 56 13 22
58 57 13 24
59 58 13 26
60 59 15 26
61 60 17 27
62 61 17 28
63 62 17 29
64 63 18 30
65 64 18 32
66 65 19 32
67 66 19 34
68 67 19 34
69 68 20 34
70 69 21 34
71 70 21 34
72 71 21 34
73 72 22 34
74 73 24 36
75 74 25 36
76 75 27 36
77 76 27 36
78 77 29 36
79 78 31 37
80 79 33 37
81 80 36 37
82 81 36 37
83 82 38 38
84 83 41 38
85 84 44 38
86 85 45 38
87 86 46 39
88 87 47 39
89 88 49 39
90 89 53 39
91 90 59 39
92 91 60 39
93 92 63 39
94 93 67 39
95 94 69 39
96 95 73 39
97 96 81 39
98 97 85 39
99 98 92 39
100 99 100 39
101 100 117 39
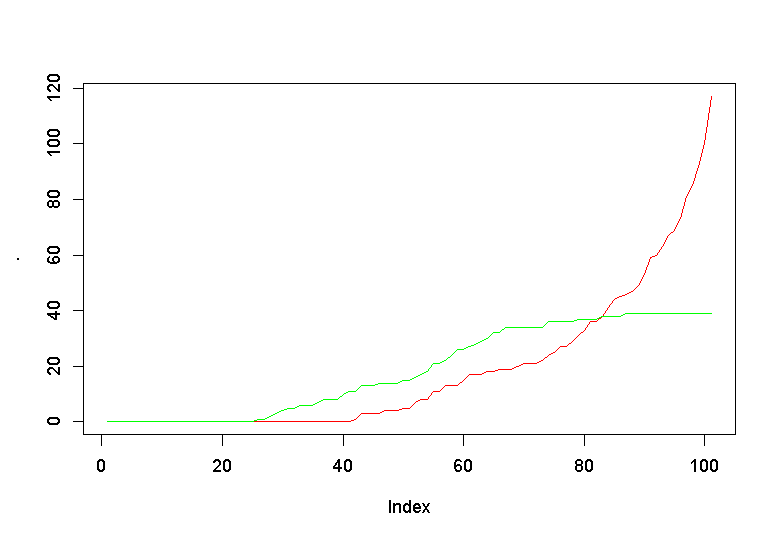
result$nb_sick %>% plot(t='l', col="red")
lines(result$nb_healhy, col="green")
CodePudding user response:
Your count is returning two rows each:
per <- 8
Valid2 %>%
select(disease_T, disease_F, predicted) %>%
filter (per >= (i/100)) %>%
count(disease_T, disease_F)
# # A tibble: 2 x 3
# disease_T disease_F n
# <lgl> <lgl> <int>
# 1 FALSE TRUE 39
# 2 TRUE FALSE 117
Further, I think your `per >= (i/100)
What I think you need is
Valid2 %>%
select(disease_T, disease_F, predicted) %>%
dplyr::filter(predicted >= (per/100)) %>%
summarize(percent = per, across(c(disease_T, disease_F), sum))
# A tibble: 1 x 3
# percent disease_T disease_F
# <dbl> <int> <int>
# 1 4 117 39
But first, a quick public-service announcement:
Iteratively adding rows to a frame using
rbind(old, newrow)works in practice but scales horribly, see "Growing Objects" in The R Inferno. for each row added, it makes a complete copy of all rows inold, which works but starts to slow down a lot. It is far better to produce a list of these new rows and thenrbindthem at one time; e.g.,out <- list(); for (...) { out <- c(out, list(newrow)); }; alldat <- do.call(rbind, out);.
Workarounds:
create an empty
out <- list()and append the results to it each time, then after the loop runbind_rowson it:plot2 <- list() for (per in 0:100) { amount <- Valid2 %>% select(disease_T, disease_F, predicted) %>% filter(predicted >= (per/100)) %>% summarize(percent = per, across(c(disease_T, disease_F), sum)) plot2 <- c(plot2, list(amount)) } plot2[1:2] # [[1]] # # A tibble: 1 x 3 # percent disease_T disease_F # <int> <int> <int> # 1 0 117 39 # [[2]] # # A tibble: 1 x 3 # percent disease_T disease_F # <int> <int> <int> # 1 1 117 39 plot2 <- bind_rows(plot2) tail(plot2, 3) # # A tibble: 6 x 3 # percent disease_T disease_F # <int> <int> <int> # 1 98 25 0 # 2 99 17 0 # 3 100 0 0use
lapplywrapped inbind_rows,plot2 <- lapply(0:100, function(per) { Valid2 %>% select(disease_T, disease_F, predicted) %>% dplyr::filter(predicted >= (per/100)) %>% summarize(percent = per, across(c(disease_T, disease_F), sum)) }) plot2 <- bind_rows(plot2)Calculate it more simply:
newout <- Valid2 %>% mutate(percent = floor(100*predicted)) %>% group_by(percent) %>% summarize(across(c(disease_T, disease_F), sum)) %>% arrange(desc(percent)) %>% mutate(across(c(disease_T, disease_F), cumsum)) head(newout, 3) # # A tibble: 3 x 3 # percent disease_T disease_F # <dbl> <int> <int> # 1 99 17 0 # 2 98 25 0 # 3 97 32 0 tail(newout, 3) # # A tibble: 3 x 3 # percent disease_T disease_F # <dbl> <int> <int> # 1 27 117 37 # 2 26 117 38 # 3 24 117 39This has a property that if a percentage did not exist previously, it is not here, but we can easily fix that with:
newout2 <- full_join(newout, tibble(percent = 0:100), by = "percent") %>% arrange(percent) %>% tidyr::fill(disease_T, disease_F, .direction = "up") %>% mutate(across(c(disease_T, disease_F), ~ coalesce(., 0L))) head(newout2, 3) # # A tibble: 3 x 3 # percent disease_T disease_F # <dbl> <int> <int> # 1 0 117 39 # 2 1 117 39 # 3 2 117 39 tail(newout2, 3) # # A tibble: 3 x 3 # percent disease_T disease_F # <dbl> <int> <int> # 1 98 25 0 # 2 99 17 0 # 3 100 0 0Which produces the same results as above:
all.equal(plot2, newout2) # [1] TRUE