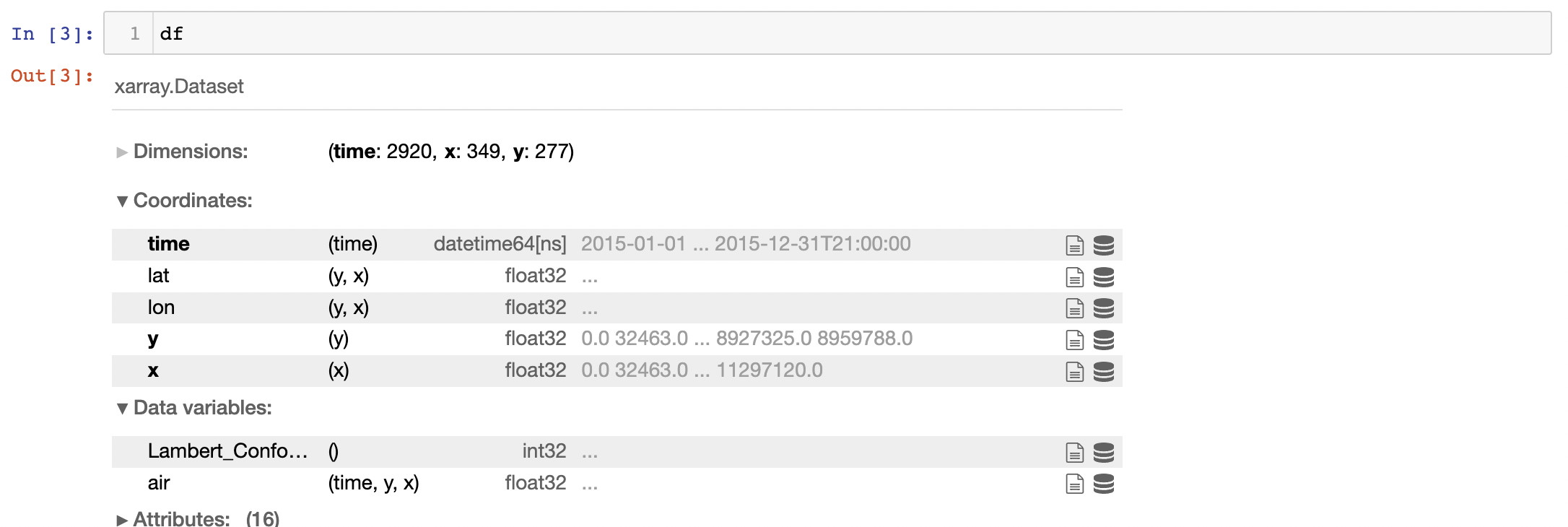
I have a gridded temperature dataset df (time: 2920 x: 349 y: 277) and a land sea mask for the same grid mf (time: 1 x: 349 y: 277) where mf.land = 1 for land grid points and mf.land = 0 for ocean points. I want to use the land sea mask to eliminate ocean points from my temperature dataset df, i.e. I only want grid points in df where mf.land = 1.
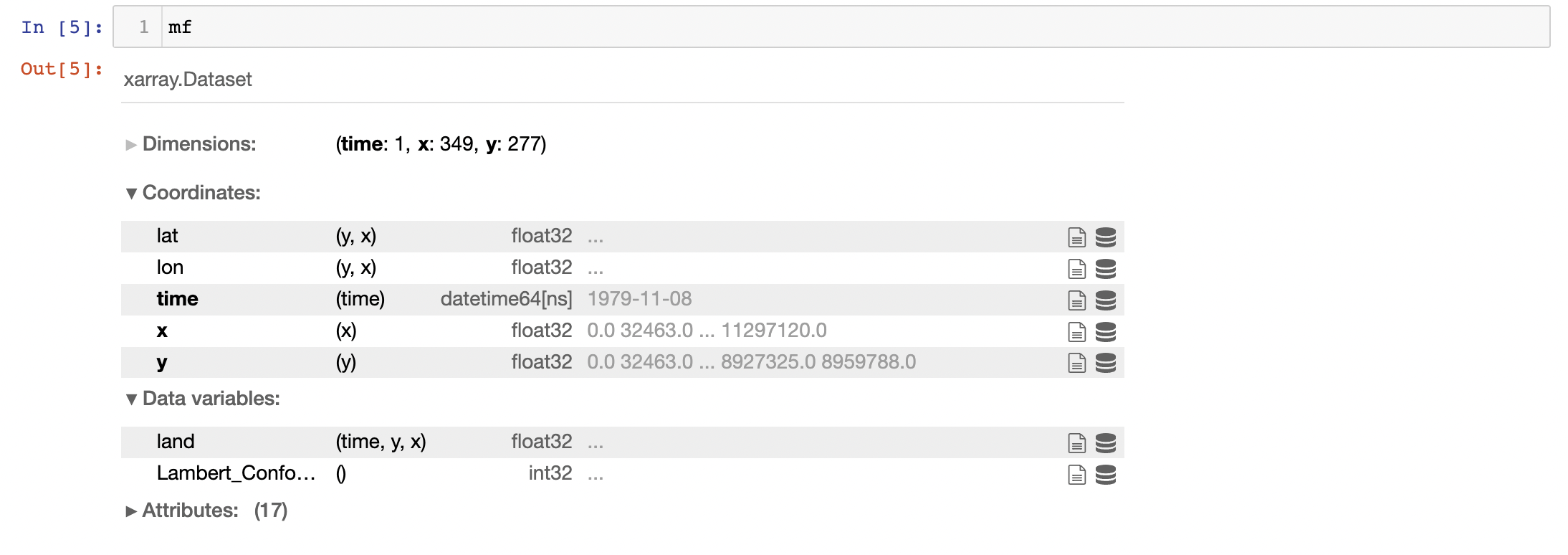
And here's what mf looks like:
I'm trying this:
#import libraries
import os
import matplotlib.pyplot as plt
from netCDF4 import Dataset as netcdf_dataset
import numpy as np
from cartopy import config
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import xarray as xr
import pandas as pd
import netCDF4 as nc
#open temperature data and land sea mask
df=xr.open_dataset('/home/mmartin/LauNath/air.2m.2015.nc')
mf=xr.open_dataset('/home/mmartin/WinterMaxThesis/NOAAGrid/land.nc')
#apply mask
mask = (mf.land >= 1)
LandOnly=df.air.loc[mask]
But am having trouble because of the difference in dimensions. How can I mask out these ocean grid points?
CodePudding user response:
The problem is actually occurring because the data arrays do have the same dimensions, but they shouldn’t. What I mean by that is that the time dimension on the land mask makes xarray think that it needs to align the two time dimensions. However, there is no overlap in the time coordinate on the two datasets, so when xarray aligns them all the data is mis-aligned in time and thus dropped. Since the land mask doesn't change through time (at least, that's what I'm assuming) it's best to exclude the time dimension from the land mask so xarray can broadcast it against the full time dimension of the data.
If you drop the time dimension on land_mask, it will broadcast as you expect:
mask = (mf.land >= 1).squeeze(['time'], drop=True)
now, you can mask your data with .where, optionally dropping all-nan slices with drop=True:
LandOnly=df.air.where(mask, drop=True)
See the user guide sections on broadcasting and automatic alignment for more info.