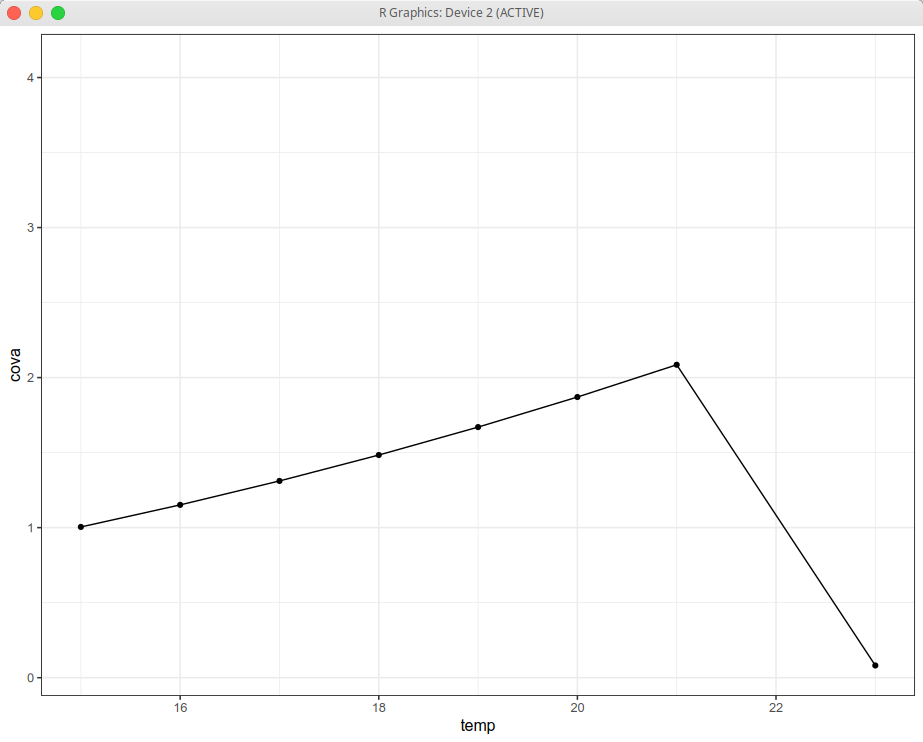
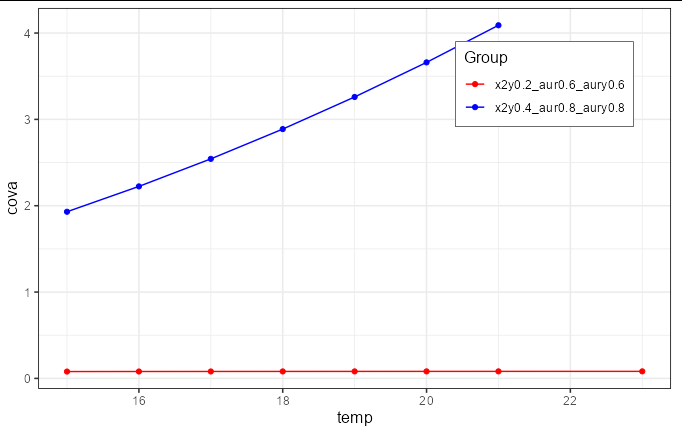
I am an old ggplot2 user, but I am getting a beating from this one. It must be something I am not noticing:
I want a simple line plot across time (x-axis) vs covariances in the y-axis. My grouping is not being picked up by ggplot2. So please consider the MWE code:
umxSim3 %>%
mutate(group = case_when(x2y == 0.4 & aur == 0.8 & aury == 0.8 ~ "x2y0.4_aur0.8_aury0.8",
x2y == 0.2 & aur == 0.6 & aury == 0.6 ~ "x2y0.2_aur0.6_aury0.6")) %>%
drop_na() %>%
ggplot(aes(temp, cova),color = as.factor(group))
geom_line(stat = "summary")
geom_point(stat = "summary")
theme_bw(12)
theme(legend.position = c(0.8, 0.8),
panel.border = element_rect(colour = "black"),
legend.background = element_rect(linetype = 1, size = 0.2, colour = 1))
scale_color_manual(values = cb_palette)
- for the data:
structure(list(temp = c(15L, 15L, 15L, 15L, 15L, 15L, 15L, 15L,
15L, 16L, 15L, 15L, 16L, 15L, 16L, 16L, 16L, 16L, 16L, 17L, 16L,
16L, 17L, 16L, 16L, 17L, 17L, 17L, 17L, 17L, 17L, 17L, 17L, 18L,
17L, 18L, 18L, 18L, 18L, 18L, 18L, 18L, 19L, 16L, 18L, 19L, 19L,
19L, 18L, 18L, 19L, 19L, 19L, 17L, 19L, 20L, 20L, 19L, 19L, 18L,
20L, 19L, 19L, 20L, 20L, 20L, 20L, 20L, 20L, 20L, 20L, 20L, 21L,
21L, 21L, 21L, 21L, 21L, 21L, 22L, 21L, 22L, 21L, 21L, 21L, 22L,
22L, 22L, 21L, 22L, 22L, 23L, 23L, 22L, 22L, 23L, 22L, 23L, 23L,
23L, 23L, 23L), immedx = c(0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02), immedy = c(0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01,
0.01, 0.01), x2y = c(0, 0, 0.2, 0, 0.2, 0.4, 0, 0.2, 0.2, 0,
0.4, 0.4, 0, 0.4, 0.2, 0.4, 0, 0.2, 0, 0.2, 0.2, 0.2, 0, 0.4,
0.4, 0, 0, 0, 0.4, 0.2, 0.4, 0.2, 0.2, 0, 0.4, 0, 0, 0.2, 0.2,
0.4, 0.4, 0, 0, 0.4, 0.2, 0.2, 0.2, 0, 0.2, 0.4, 0, 0.4, 0, 0.4,
0.2, 0, 0, 0.4, 0.2, 0.4, 0.2, 0.4, 0.4, 0, 0.2, 0.4, 0, 0.2,
0.2, 0.4, 0.4, 0.4, 0, 0.2, 0, 0.2, 0.4, 0, 0, 0, 0.2, 0, 0.4,
0.4, 0.2, 0, 0.4, 0, 0.4, 0.2, 0.2, 0, 0.2, 0.2, 0.4, 0, 0.4,
0, 0.4, 0, 0.2, 0.4), aur = c(0.6, 0.6, 0.6, 0.8, 0.6, 0.6, 0.8,
0.8, 0.8, 0.6, 0.8, 0.6, 0.8, 0.8, 0.6, 0.6, 0.6, 0.8, 0.8, 0.6,
0.8, 0.6, 0.6, 0.8, 0.6, 0.8, 0.6, 0.8, 0.6, 0.8, 0.6, 0.6, 0.8,
0.6, 0.8, 0.6, 0.8, 0.8, 0.6, 0.6, 0.6, 0.8, 0.6, 0.8, 0.6, 0.6,
0.6, 0.8, 0.8, 0.8, 0.6, 0.6, 0.8, 0.8, 0.8, 0.6, 0.6, 0.6, 0.8,
0.8, 0.6, 0.8, 0.8, 0.8, 0.8, 0.6, 0.8, 0.6, 0.8, 0.6, 0.8, 0.8,
0.6, 0.6, 0.6, 0.6, 0.6, 0.8, 0.8, 0.6, 0.8, 0.6, 0.8, 0.6, 0.8,
0.8, 0.6, 0.8, 0.8, 0.6, 0.8, 0.6, 0.6, 0.8, 0.6, 0.6, 0.8, 0.8,
0.6, 0.8, 0.6, 0.6), aury = c(0.8, 0.6, 0.6, 0.6, 0.8, 0.6, 0.8,
0.6, 0.8, 0.6, 0.6, 0.8, 0.6, 0.8, 0.6, 0.6, 0.8, 0.6, 0.8, 0.6,
0.8, 0.8, 0.8, 0.6, 0.8, 0.6, 0.6, 0.8, 0.8, 0.6, 0.6, 0.8, 0.8,
0.8, 0.6, 0.6, 0.6, 0.6, 0.6, 0.8, 0.6, 0.8, 0.6, 0.8, 0.8, 0.6,
0.8, 0.6, 0.8, 0.6, 0.8, 0.6, 0.8, 0.8, 0.6, 0.8, 0.6, 0.8, 0.8,
0.8, 0.6, 0.6, 0.8, 0.6, 0.6, 0.6, 0.8, 0.8, 0.8, 0.8, 0.8, 0.6,
0.6, 0.6, 0.8, 0.8, 0.6, 0.6, 0.8, 0.6, 0.6, 0.8, 0.6, 0.8, 0.8,
0.6, 0.6, 0.8, 0.8, 0.8, 0.6, 0.6, 0.6, 0.8, 0.8, 0.8, 0.6, 0.6,
0.6, 0.8, 0.8, 0.8), cova = c(0.0767650449455536, 0.023479773134789,
0.079250541929235, 0.0497028270157154, 0.217607799079627, 0.168683335594918,
0.150794456586447, 0.27283026101063, 0.690314230152603, 0.0235259541982523,
0.702603056392517, 0.490115926773996, 0.0505076752505145, 1.93010433620619,
0.0798575573537466, 0.171573575141921, 0.0784006741340135, 0.28486748087407,
0.156814170306613, 0.0803151708822509, 0.754445007375547, 0.226801448778645,
0.079735730985492, 0.755994056890623, 0.525290423561159, 0.0511604604964653,
0.0235558887825711, 0.162030264083756, 0.559246976325596, 0.295696528479825,
0.173935711302792, 0.235063054293184, 0.818154209385252, 0.080823259527356,
0.80766448894273, 0.0235752474868944, 0.0516895405415864, 0.305415501682546,
0.0806599420516001, 0.59193655065581, 0.175863895489332, 0.166532200794381,
0.0235877421429864, 2.22373977593273, 0.242470710342392, 0.0809195781739381,
0.249100119500662, 0.0521181251083318, 0.881239650574519, 0.857515819530478,
0.0817077551411662, 0.177436314599814, 0.170404901664453, 2.54251804738705,
0.314120482284585, 0.0824262284334331, 0.0235957929692925, 0.62332889414795,
0.943529017271466, 2.88745078017984, 0.0811150355928273, 0.905481581223059,
3.25958506793805, 0.0524651556999694, 0.321903676726418, 0.178717602990971,
0.173727014344408, 0.255023388431499, 1.00487624869021, 0.653409404225594,
3.66000427224439, 0.951522116145514, 0.0236009729296943, 0.0812621413155391,
0.0830092693966824, 0.260308295568005, 0.179761001622845, 0.0527460575456124,
0.17657007247267, 0.0236043015726037, 0.328852247070274, 0.0834820393132251,
0.995620065250206, 0.682176435239891, 1.06515843598909, 0.0529733721858174,
0.180610243434384, 0.178998254606776, 4.08982898920573, 0.265017899614053,
0.335047641163975, 0.0236064382275949, 0.0814561191768848, 1.12427313098231,
0.709638986736915, 0.0838651574035433, 1.03777649778554, 0.0531572840695336,
0.181301171395821, 0.181068531268503, 0.269210391598863, 0.735814722588949
), ncp = c(0.00000046267086872831, 0.0000000313011696562171,
-2364.79037081984, -0.000000597414327785373, 351.394396380274,
151.990268357971, 0.00000743399141356349, 19.7703358054714, 2440.01464180378,
-7240.58302952931, 70.9914281773526, 149.740843244857, -3309.66286159754,
16647.7277528127, -2582.00822814501, -1561.43493972577, -0.000000475047272630036,
19.6414650627339, 0.0000108720123535022, 315.029701597647, 135.427727860704,
22.4076952430914, 0.0000000233412720263004, -752.562054366463,
80.5575949394988, -0.00000442085001850501, -1179.58693431943,
0.000000114778231363744, -7327.08611100476, -1949.39119122034,
-2388.61869873563, 22.1016658367153, 2824.56886877323, -0.0000042213941924274,
-730.911226568125, -1099.31476127236, 0.00000000339059624820948,
2378.51963226983, -3114.53016945277, 3984.12834500313, 175.787574969581,
0.00000653910683467984, -0.000000110070686787367, 1181.21838523608,
21.8320325434906, -3186.31096406218, 345.549440360701, 13365.6075074154,
14.6078058980493, 2258.65831825051, -700.956671146072, -406.573908048507,
0.0000124756261357106, 1164.89840225065, 19.3309273536142, -0.00000834093225421384,
-6412.21120826376, 69.809013458027, 1435.53987527536, 1438.76661482502,
28.9054297961484, 2385.37075061088, 27660.1530534023, 0.000000160784111358225,
-3822.79946768601, -2315.2593892417, 0.00000594364973949268,
27.1325407072363, 3411.75060408833, 2138.85256412776, 30763.6924561956,
64.9265390225191, -2428.93211152896, -2914.7903949441, 0.00000096066651167348,
678.510061382112, -826.474972046213, 0.000000630076101515442,
0.00000610958522884175, -0.00000443263706983998, -3691.07725347468,
0.00000263373658526689, 12496.6406096131, 111.59293522677, 13.801709866937,
0.0000000575091689825058, -482.465671315367, 0.00000894447293831035,
-944.987300472989, 26.6739735744122, -1686.73710901346, 0.000000106025254353881,
-1838.70931961411, 1602.34840365657, -5004.48179096662, -0.0000282387336483225,
1476.99946196973, 0.000000139494659379125, -3032.83538644376,
0.000000328800524584949, -4550.10670159884, 2268.41146332065),
df = c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
proximal = c(0.02, 0.02, -0.039, 0.02, 0.02, 0.023, 0.02,
0.02, 0.02, -0.229, 0.02, 0.02, 0.096, 0.02, -0.241, 0.174,
0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.04, 0.02, 0.02,
0.022, 0.02, 0.129, -0.093, -0.216, 0.02, 0.02, 0.02, -0.061,
0.014, 0.02, -0.025, -0.715, 0.024, 0.02, 0.02, 0.02, 0.02,
0.02, 0.036, 0.02, -6.749, 0.02, 0.02, 0.012, -0.032, 0.02,
0.02, 0.02, 0.02, -0.093, 0.021, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, -0.27, -0.194, 0.02, 0.02, 0.02, 0.02, 0.02,
0.022, -0.071, -0.103, 0.02, 0.02, 0.121, 0.02, 0.02, 0.02,
0.083, 0.02, 0.02, -0.013, 0.02, 0.02, -0.401, 0.02, 0.115,
0.02, 0.084, 0.02, 0.041, 0.018, -0.099, 0.02, 0.02, 0.02,
-0.089, 0.02, -0.056, -0.063), distal = c(0, 0, 0.209, 0,
0.2, 0.396, 0, 0.2, 0.2, 0.145, 0.4, 0.4, -0.09, 0.4, 0.341,
0.273, 0, 0.2, 0, 0.2, 0.2, 0.2, 0, 0.392, 0.4, 0, -0.019,
0, 0.503, 0.29, 0.54, 0.2, 0.2, 0, 0.474, 0, 0, 0.237, 0.621,
0.396, 0.4, 0, 0, 0.4, 0.2, 0.195, 0.2, 6.526, 0.2, 0.4,
0.005, 0.425, 0, 0.4, 0.2, 0, 0.143, 0.4, 0.2, 0.4, 0.2,
0.4, 0.4, 0, 0.512, 0.613, 0, 0.2, 0.2, 0.4, 0.4, 0.396,
0.085, 0.264, 0, 0.2, 0.309, 0, 0, 0, 0.173, 0, 0.4, 0.409,
0.2, 0, 0.708, 0, 0.307, 0.2, 0.131, 0, 0.205, 0.201, 0.594,
0, 0.4, 0, 0.519, 0, 0.238, 0.465), historical = c(0.02,
0.02, 0.02, 0.02, 0.02, 0.026, 0.02, 0.02, 0.02, 0.062, 0.02,
0.02, -0.15, 0.02, -0.225, -0.121, 0.02, 0.02, 0.02, 0.02,
0.02, 0.02, 0.02, 0.085, 0.02, 0.02, 0.037, 0.02, 0.124,
-0.082, -0.202, 0.02, 0.02, 0.02, 0.07, 0.024, 0.02, -0.022,
-0.69, 0.023, 0.02, 0.02, 0.02, 0.02, 0.02, 0.038, 0.02,
0.151, 0.02, 0.02, 0.082, -0.063, 0.02, 0.02, 0.02, 0.02,
0.159, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, 0.02, -0.117,
-0.033, 0.02, 0.02, 0.02, 0.02, 0.02, 0.025, -0.086, -0.044,
0.02, 0.022, -0.168, 0.02, 0.02, 0.02, 0.299, 0.02, 0.02,
-0.015, 0.02, 0.02, -0.128, 0.02, -0.088, 0.02, 0.035, 0.02,
0.041, 0.019, -0.218, 0.02, 0.02, 0.02, -0.062, 0.02, -0.088,
-0.057)), class = "data.frame", row.names = c(NA, -102L))
For this data I am getting:
Whereas I expected it to be colored by two groups:
- 2y == 0.4 & aur == 0.8 & aury == 0.8 ~ "x2y0.4_aur0.8_aury0.8",
- x2y == 0.2 & aur == 0.6 & aury == 0.6 ~ "x2y0.2_aur0.6_aury0.6")
CodePudding user response:
You need to move the color specification inside the aes call:
umxSim3 %>%
mutate(group = case_when(x2y == 0.4 & aur == 0.8 & aury == 0.8 ~ "x2y0.4_aur0.8_aury0.8",
x2y == 0.2 & aur == 0.6 & aury == 0.6 ~ "x2y0.2_aur0.6_aury0.6")) %>%
drop_na() %>%
ggplot(aes(temp, cova, color = as.factor(group)))
geom_line(stat = "summary")
geom_point(stat = "summary")
theme_bw(12)
theme(legend.position = c(0.8, 0.8),
panel.border = element_rect(colour = "black"),
legend.background = element_rect(linetype = 1, size = 0.2, colour = 1))
scale_color_manual(values = c('red', 'blue'), name = 'Group')
Note you didn't provide the cb_palette object, so I have just set the colors to red and blue for this example.