can someone help me with the construction of the meta-analysis sub-plot with the ggplot2 library.
Here is the reproducible sample:
library(ggplot2)
library(meta)
study <- c("Study 1","Study 2","Study 3","Study 4","Study 5",
"Study 6","Study 7","Study 8","Study 9","Study 10",
"Study 11","Study 12","Study 13","Study 14","Study 15")
nT <- c(155,31,75,18,8,57,34,110,60,20,11,32,36,97,80)
meanT <- c(55,27,64,66,14,19,52,21,30,45,32,80,40,25,70)
sdT <- c(47,7,17,20,8,7,45,16,27,11,6,22,31,4,32)
nC <- c(156,32,71,18,13,52,33,183,52,22,14,32,44,93,81)
meanC <- c(75,29,119,137,18,18,41,31,23,16,44,65,22,11,90)
sdC <- c(64,4,29,48,11,4,34,27,20,5,21,37,8,2,55)
pH <- c("high","low","high","low","low","high","low","high",
"high","low","low","low","low","high","high")
dt <- data.frame(study,nT,meanT,sdT,nC,meanC,sdC,pH)
head(dt, 5)
# meta-analysis model
dt$pH <- factor(dt$pH, levels = c("low", "high"))
m1 <- metacont(nT, meanT, sdT,
nC, meanC, sdC,
fixed = FALSE,
random = TRUE,
subgroup = pH,
data = dt)
summary(m1)
# forest plot with two pH levels (and individual studies)
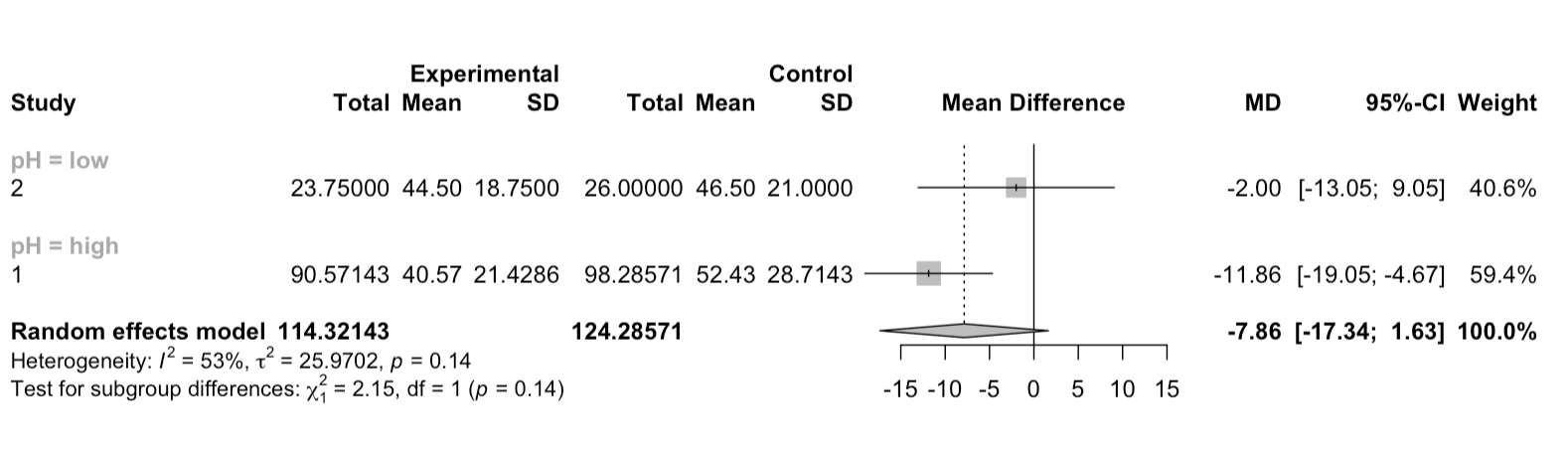
forest(m1, subgroup = TRUE, print.byvar = FALSE)
I want to see the plot with the effect sizes (and CIs) only for two pH levels without individual studies.
Many thanks in advance!
CodePudding user response:
What you could do is create a dataframe which is a group_by of your pH where you calculate the mean per columns, so you have two rows which describe the low and high pH like this:
library(ggplot2)
library(dplyr)
library(meta)
study <- c("Study 1","Study 2","Study 3","Study 4","Study 5",
"Study 6","Study 7","Study 8","Study 9","Study 10",
"Study 11","Study 12","Study 13","Study 14","Study 15")
nT <- c(155,31,75,18,8,57,34,110,60,20,11,32,36,97,80)
meanT <- c(55,27,64,66,14,19,52,21,30,45,32,80,40,25,70)
sdT <- c(47,7,17,20,8,7,45,16,27,11,6,22,31,4,32)
nC <- c(156,32,71,18,13,52,33,183,52,22,14,32,44,93,81)
meanC <- c(75,29,119,137,18,18,41,31,23,16,44,65,22,11,90)
sdC <- c(64,4,29,48,11,4,34,27,20,5,21,37,8,2,55)
pH <- c("high","low","high","low","low","high","low","high",
"high","low","low","low","low","high","high")
dt <- data.frame(study,nT,meanT,sdT,nC,meanC,sdC,pH)
head(dt, 5)
#> study nT meanT sdT nC meanC sdC pH
#> 1 Study 1 155 55 47 156 75 64 high
#> 2 Study 2 31 27 7 32 29 4 low
#> 3 Study 3 75 64 17 71 119 29 high
#> 4 Study 4 18 66 20 18 137 48 low
#> 5 Study 5 8 14 8 13 18 11 low
# This part to aggregated all colums
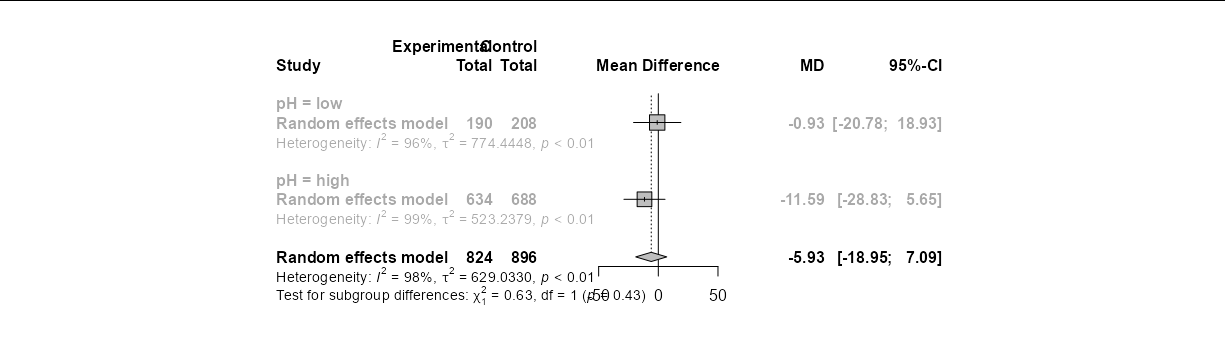
dt <- dt %>%
group_by(pH) %>%
summarise(across(everything(), mean))
# meta-analysis model
dt$pH <- factor(dt$pH, levels = c("low", "high"))
m1 <- metacont(nT, meanT, sdT,
nC, meanC, sdC,
fixed = FALSE,
random = TRUE,
subgroup = pH,
data = dt)
# forest plot with two pH levels (and individual studies)
forest(m1, subgroup = TRUE, print.byvar = FALSE)
I guess this function simply has too many parameters to make it easy to find the right options...