I have two data frames, d1 and d2:
d1 <- data.frame(ID=c("x1", "x1", "x1", "x1", "x1", "x2", "x2", "x2", "x2", "x2", "x2", "x2"),
stat=c("A", "B", "C", "D", "E", "H", "I", "J", "J", "K", "L", "M"),
OD=c("AE", "AE", "AE", "AE", "AE", "HJ", "HJ", "HJ", "JM", "JM", "JM", "JM"))
d2 <- data.frame(ID=c("x1", "x1", "x1", "x2", "x2", "x2"),
OD=c("AE", "AE", "BD", "HJ", "HJ", "JM"),
prod=c("p_2", "p_1", "p_3", "p_5", "p_4", "p_5"),
wgh=c(1000, 1300, 300, 2300, 1800, 2300))
The data is grouped by 'ID'. The 'stat' column are stations (or sites) visited. The 'OD' column is the origin and destination stations concatenated.
d2 has product id:s 'prod' and their weight 'wgh', which I want to join to d1. The variables to join by are ID and OD.
d1
ID stat OD
1 x1 A AE
2 x1 B AE
3 x1 C AE
4 x1 D AE
5 x1 E AE
6 x2 H HJ
7 x2 I HJ
8 x2 J HJ
9 x2 J JM
10 x2 K JM
11 x2 L JM
12 x2 M JM
d2
ID OD prod wgh
1 x1 AE p_2 1000
2 x1 AE p_1 1300
3 x1 BD p_3 300 # origin B & and dest. D within the range of 'stat' in d1, but 'OD' doesn't match
4 x2 HJ p_5 2300
5 x2 HJ p_4 1800
6 x2 JM p_5 2300
Sometimes OD matches between the two data sets, e.g. ID = "x1" & OD = "AE". Fine.
However, the main issue is that there are cases in d2 when the origin occurs after the origin in d1, and when the destination occurs before the destination in d1. For example, in d2, where ID = x1, the OD = BD is not present in d1 (comment in data). The stations in d2, B to D, have been visited on the way between A and E. This means that 'prod' and 'wgh' from d2 should be joined to rows in d1 where 'stat' is from B to D.
In the result, the number of rows of d1, should remain the same. However, the number of columns would be increased, as I was thinking to put up the available prod product ids as variables, and then match the wgh weights to those.
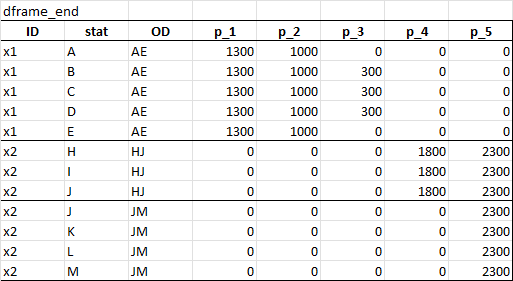
The result table would look something like this:
Maybe merge and/or dcast is a good way to start. I was even thinking of ignoring OD and just make an origin and destination column, then somehow fill down values in between when they match with stat, but I got stuck.
Note that my real data are millions of row long.
CodePudding user response:
A suggestion for the reshaping
library(tidyverse)
dframe_2 %>%
pivot_wider(names_from = prod,
values_from = wgh,
values_fill = 0)
left_join(dframe_1, dframe_2)
ID stat OD p_18 p_15 p_3 p_16 p_14
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 X1 A AE 1000 1300 0 0 0
2 X1 B AE 1000 1300 0 0 0
3 X1 C AE 1000 1300 0 0 0
4 X1 D AE 1000 1300 0 0 0
5 X1 E AE 1000 1300 0 0 0
6 X2 F HJ 0 0 0 2300 1800
7 X2 G HJ 0 0 0 2300 1800
8 X2 H HJ 0 0 0 2300 1800
9 X2 I JM 0 0 0 2300 0
10 X2 J JM 0 0 0 2300 0
11 X2 K JM 0 0 0 2300 0
12 X2 L JM 0 0 0 2300 0
CodePudding user response:
You want to expand your AE to multiple entries, alphabetically - you can use the LETTERS built-in for a reference alphabet, and use match to find the location of the initial and final letter. For example
OD <- "AE"
ODarray <- LETTERS[match(substr(OD,1,1), LETTERS) : match(substr(OD,2,2), LETTERS)]
print(ODarray)
[1] "A" "B" "C" "D" "E"
Then, in order to link your d1 and d2, you can loop this process to create a bridge df between OD and stat:
bridge = do.call(rbind, lapply(unique(d2$OD), function(od)
data.frame("OD" = od,
"stat" = LETTERS[match(substr(od,1,1), LETTERS) :
match(substr(od,2,2), LETTERS)]
))
print(bridge)
OD stat
1 AE A
2 AE B
3 AE C
4 AE D
5 AE E
6 BD B
7 BD C
8 BD D
9 HJ H
10 HJ I
11 HJ J
12 JM J
13 JM K
14 JM L
15 JM M
Edit: the next problem is that you want to match the OD in d1 to a different OD in d2, for example the values in d2 that have an OD = "BD" should actually be matched to the OD = "AE" in d1.
Since your letter ranges overlap (e.g. HJ and JM), you will have to design a matching array manually (if you have hundreds of combinations, there's also a regex method, but I am assuming here that there are only a handful of possible values of d2$OD that are not in d1$OD already)
The easiest method is to create a named array a lookup, where each value in d1$OD maps to itself, and values that are in d2$OD but not d1$OD have an explicitly defined translation:
ODlookup <- c(
setNames(unique(d1$OD), unique(d1$OD)), # identity map
"BD" = "AE") # manual translations
d2$newOD <- ODlookup[d2$OD]
Merge the bridge into d2, and the resulting data.frame into d1. You'll have to specify you want to use the newly-created d2$newOD. Done!
merge(d1,merge(d2, bridge),
by.x = c("ID", "OD", "stat"),
by.y = c("ID", "newOD", "stat"))
ID OD stat OD.y prod wgh
1 x1 AE A AE p_2 1000
2 x1 AE A AE p_1 1300
3 x1 AE B AE p_1 1300
4 x1 AE B AE p_2 1000
5 x1 AE B BD p_3 300
6 x1 AE C AE p_1 1300
7 x1 AE C BD p_3 300
8 x1 AE C AE p_2 1000
9 x1 AE D AE p_1 1300
10 x1 AE D BD p_3 300
11 x1 AE D AE p_2 1000
12 x1 AE E AE p_1 1300
13 x1 AE E AE p_2 1000
14 x2 HJ H HJ p_5 2300
15 x2 HJ H HJ p_4 1800
16 x2 HJ I HJ p_5 2300
17 x2 HJ I HJ p_4 1800
18 x2 HJ J HJ p_5 2300
19 x2 HJ J HJ p_4 1800
20 x2 JM J JM p_5 2300
21 x2 JM K JM p_5 2300
22 x2 JM L JM p_5 2300
23 x2 JM M JM p_5 2300
You can then follow Tom Hoel's answer for the reshaping (although consider whether this long format may actually be more useful, instead of creating values of 0 where you are missing data)